7T coil comparison: processing of phantom data#
Setup environment#
# Imports
from datetime import datetime, timedelta
import os
import json
import glob
import matplotlib.pyplot as plt
import numpy as np
import nibabel as nib
import pandas as pd
from scipy.stats import iqr
import shutil
import zipfile
from scipy.interpolate import interp1d
from scipy.ndimage import uniform_filter1d
from pathlib import Path
# Define constants
GAMMA = 2.675e8; # [rad / (s T)]
%matplotlib inline
start_time = datetime.now()
# Download data from OpenNeuro ⏳
!openneuro-py download --dataset ds005090 --target-dir data-phantom/
👋 Hello! This is openneuro-py 2024.2.0. Great to see you! 🤗
👉 Please report problems 🤯 and bugs 🪲 at
https://github.com/hoechenberger/openneuro-py/issues
🌍 Preparing to download ds005090 …
📁 Traversing directories for ds005090 : 0 entities [00:00, ? entities/s]
📁 Traversing directories for ds005090 : 5 entities [00:00, 14.66 entities/s]
📁 Traversing directories for ds005090 : 45 entities [00:00, 64.80 entities/s]
📁 Traversing directories for ds005090 : 105 entities [00:01, 117.24 entities/s]
📁 Traversing directories for ds005090 : 147 entities [00:01, 147.16 entities/s]
📁 Traversing directories for ds005090 : 169 entities [00:01, 154.41 entities/s]
📁 Traversing directories for ds005090 : 219 entities [00:01, 195.38 entities/s]
📁 Traversing directories for ds005090 : 241 entities [00:01, 194.36 entities/s]
📁 Traversing directories for ds005090 : 307 entities [00:01, 264.58 entities/s]
📁 Traversing directories for ds005090 : 379 entities [00:01, 322.70 entities/s]
📁 Traversing directories for ds005090 : 418 entities [00:02, 208.36 entities/s]
📥 Retrieving up to 418 files (5 concurrent downloads).
participants.tsv: 0%| | 0.00/313 [00:00<?, ?B/s]
.bidsignore: 0%| | 0.00/92.0 [00:00<?, ?B/s]
sub-CRMBM_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
CHANGES: 0%| | 0.00/91.0 [00:00<?, ?B/s]
dataset_description.json: 0%| | 0.00/314 [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined1_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/737k [00:00<?, ?B/s]
sub-CRMBM_T2starw.nii.gz: 0%| | 0.00/864k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined2_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined2_T2starw.nii.gz: 11%| | 83.6k/737k [00:00<00:01, 623kB
sub-CRMBM_T2starw.nii.gz: 10%|█▍ | 85.2k/864k [00:00<00:01, 637kB/s]
sub-CRMBM_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/676k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined3_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined2_T2starw.nii.gz: 48%|▍| 356k/737k [00:00<00:00, 1.48MB
sub-CRMBM_T2starw.nii.gz: 41%|█████▊ | 357k/864k [00:00<00:00, 1.49MB/s]
sub-CRMBM_rec-uncombined1_T2starw.nii.gz: 12%| | 83.6k/676k [00:00<00:01, 570kB
sub-CRMBM_rec-uncombined5_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined1_T2starw.nii.gz: 53%|▌| 356k/676k [00:00<00:00, 1.44MB
sub-CRMBM_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/799k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined4_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/700k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined3_T2starw.nii.gz: 10%| | 78.0k/799k [00:00<00:01, 568kB
sub-CRMBM_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/707k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined3_T2starw.nii.gz: 44%|▍| 350k/799k [00:00<00:00, 1.68MB
sub-CRMBM_rec-uncombined4_T2starw.nii.gz: 12%| | 83.6k/700k [00:00<00:01, 613kB
sub-CRMBM_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/606k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined5_T2starw.nii.gz: 12%| | 83.6k/707k [00:00<00:01, 612kB
sub-CRMBM_rec-uncombined6_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined3_T2starw.nii.gz: 93%|▉| 740k/799k [00:00<00:00, 2.36MB
sub-CRMBM_rec-uncombined5_T2starw.nii.gz: 43%|▍| 305k/707k [00:00<00:00, 1.32MB
sub-CRMBM_rec-uncombined4_T2starw.nii.gz: 48%|▍| 339k/700k [00:00<00:00, 1.25MB
sub-CRMBM_rec-uncombined7_T2starw.nii.gz: 14%|▏| 83.6k/606k [00:00<00:00, 562kB
sub-CRMBM_rec-uncombined7_T2starw.nii.gz: 56%|▌| 339k/606k [00:00<00:00, 1.37MB
sub-CRMBM_rec-uncombined7_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/677k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined8_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM_acq-anat_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/639k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined6_T2starw.nii.gz: 13%|▏| 85.2k/677k [00:00<00:00, 643kB
sub-CRMBM_rec-uncombined8_T2starw.nii.gz: 12%| | 77.0k/639k [00:00<00:01, 568kB
sub-CRMBM_rec-uncombined6_T2starw.nii.gz: 53%|▌| 357k/677k [00:00<00:00, 1.51MB
sub-CRMBM_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/833k [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-CRMBM_rec-uncombined8_T2starw.nii.gz: 57%|▌| 366k/639k [00:00<00:00, 1.47MB
sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.88M [00:00<?, ?B/s]
sub-CRMBM_acq-anat_TB1TFL.nii.gz: 10%|▌ | 83.6k/833k [00:00<00:01, 489kB/s]
sub-CRMBM_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.5k/3.88M [00:00<00:06, 598kB
sub-CRMBM_acq-anat_TB1TFL.nii.gz: 35%|██ | 288k/833k [00:00<00:00, 1.02MB/s]
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz: 10%| | 407k/3.88M [00:00<00:02, 1.63MB
sub-CRMBM_acq-anat_TB1TFL.nii.gz: 67%|████ | 560k/833k [00:00<00:00, 1.43MB/s]
sub-CRMBM_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.65M [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz: 25%|▏| 985k/3.88M [00:00<00:00, 3.16MB
sub-CRMBM_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/11.7M [00:00<00:22, 553kB/
sub-CRMBM_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.65M [00:00<00:04, 5
sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz: 77%|▊| 2.99M/3.88M [00:00<00:00, 9.16M
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 3%| | 356k/11.7M [00:00<00:08, 1.45MB/
sub-CRMBM_acq-coilQaSagSmall_GFactor.nii.gz: 14%|▏| 373k/2.65M [00:00<00:01, 1.
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 12%| | 1.36M/11.7M [00:00<00:02, 4.92MB
sub-CRMBM_acq-coilQaSagSmall_GFactor.nii.gz: 50%|▍| 1.31M/2.65M [00:00<00:00, 4
sub-CRMBM_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-CRMBM_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/90.7k [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaSagSmall_GFactor.nii.gz: 100%|▉| 2.64M/2.65M [00:00<00:00, 7
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 25%|▎| 2.95M/11.7M [00:00<00:01, 8.71MB
sub-CRMBM_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 49%|▍| 5.76M/11.7M [00:00<00:00, 15.2MB
sub-CRMBM_acq-famp-0.66_TB1DREAM.nii.gz: 94%|▉| 85.2k/90.7k [00:00<00:00, 631kB
sub-CRMBM_acq-coilQaTra_GFactor.nii.gz: 85%|▊| 9.93M/11.7M [00:00<00:00, 24.1MB
sub-CRMBM_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.4k [00:00<?, ?B/s]
sub-CRMBM_acq-famp_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-CRMBM_acq-famp_TB1TFL.json: 0%| | 0.00/2.69k [00:00<?, ?B/s]
sub-CRMBM_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 85.2k/91.4k [00:00<00:00, 598kB/
sub-CRMBM_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/90.9k [00:00<?, ?B/s]
sub-CRMBM_acq-famp_TB1DREAM.nii.gz: 94%|██▊| 85.2k/90.9k [00:00<00:00, 620kB/s]
sub-CRMBM_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/919k [00:00<?, ?B/s]
sub-CRMBM_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/78.5k [00:00<?, ?B/s]
sub-CRMBM_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/919k [00:00<00:01, 695kB/s]
sub-CRMBM_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/85.8k [00:00<?, ?B/s]
sub-CRMBM_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 78.5k/78.5k [00:00<00:00, 628kB
sub-CRMBM_acq-famp_TB1TFL.nii.gz: 39%|██▎ | 356k/919k [00:00<00:00, 1.48MB/s]
sub-CRMBM_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM_acq-refv-1.5_TB1DREAM.nii.gz: 90%|▉| 77.0k/85.8k [00:00<00:00, 504kB/
sub-CRMBM_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/82.4k [00:00<?, ?B/s]
sub-CRMBM_acq-refv_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM_acq-refv_TB1DREAM.nii.gz: 100%|███| 82.4k/82.4k [00:00<00:00, 565kB/s]
sub-MGH_rec-uncombined01_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MGH_T2starw.nii.gz: 0%| | 0.00/1.14M [00:00<?, ?B/s]
sub-MGH_T2starw.nii.gz: 7%|█ | 85.2k/1.14M [00:00<00:01, 626kB/s]
sub-MGH_rec-uncombined02_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/804k [00:00<?, ?B/s]
sub-MGH_T2starw.nii.gz: 31%|████▌ | 357k/1.14M [00:00<00:00, 1.43MB/s]
sub-MGH_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/717k [00:00<?, ?B/s]
sub-MGH_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/718k [00:00<?, ?B/s]
sub-MGH_rec-uncombined01_T2starw.nii.gz: 10%| | 83.6k/804k [00:00<00:01, 613kB/
sub-MGH_rec-uncombined02_T2starw.nii.gz: 12%| | 85.2k/717k [00:00<00:01, 629kB/
sub-MGH_rec-uncombined03_T2starw.nii.gz: 12%| | 85.2k/718k [00:00<00:01, 629kB/
sub-MGH_rec-uncombined01_T2starw.nii.gz: 44%|▍| 356k/804k [00:00<00:00, 1.42MB/
sub-MGH_rec-uncombined03_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined02_T2starw.nii.gz: 50%|▍| 357k/717k [00:00<00:00, 1.29MB/
sub-MGH_rec-uncombined03_T2starw.nii.gz: 47%|▍| 340k/718k [00:00<00:00, 1.25MB/
sub-MGH_rec-uncombined01_T2starw.nii.gz: 100%|█| 804k/804k [00:00<00:00, 2.50MB/
sub-MGH_rec-uncombined04_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined05_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/725k [00:00<?, ?B/s]
sub-MGH_rec-uncombined06_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/798k [00:00<?, ?B/s]
sub-MGH_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/794k [00:00<?, ?B/s]
sub-MGH_rec-uncombined04_T2starw.nii.gz: 12%| | 83.6k/725k [00:00<00:01, 613kB/
sub-MGH_rec-uncombined05_T2starw.nii.gz: 11%| | 85.2k/798k [00:00<00:01, 622kB/
sub-MGH_rec-uncombined07_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined06_T2starw.nii.gz: 12%| | 94.0k/794k [00:00<00:01, 640kB/
sub-MGH_rec-uncombined04_T2starw.nii.gz: 49%|▍| 356k/725k [00:00<00:00, 1.42MB/
sub-MGH_rec-uncombined05_T2starw.nii.gz: 51%|▌| 408k/798k [00:00<00:00, 1.60MB/
sub-MGH_rec-uncombined06_T2starw.nii.gz: 40%|▍| 315k/794k [00:00<00:00, 1.14MB/
sub-MGH_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/773k [00:00<?, ?B/s]
sub-MGH_rec-uncombined08_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined07_T2starw.nii.gz: 11%| | 83.6k/773k [00:00<00:01, 549kB/
sub-MGH_rec-uncombined07_T2starw.nii.gz: 46%|▍| 356k/773k [00:00<00:00, 1.57MB/
sub-MGH_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/853k [00:00<?, ?B/s]
sub-MGH_rec-uncombined10_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined09_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined08_T2starw.nii.gz: 8%| | 66.6k/853k [00:00<00:01, 669kB/
sub-MGH_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/805k [00:00<?, ?B/s]
sub-MGH_rec-uncombined08_T2starw.nii.gz: 20%|▏| 169k/853k [00:00<00:00, 845kB/s
sub-MGH_rec-uncombined09_T2starw.nii.gz: 10%| | 83.6k/805k [00:00<00:01, 613kB/
sub-MGH_rec-uncombined11_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/838k [00:00<?, ?B/s]
sub-MGH_rec-uncombined08_T2starw.nii.gz: 84%|▊| 713k/853k [00:00<00:00, 2.26MB/
sub-MGH_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/857k [00:00<?, ?B/s]
sub-MGH_rec-uncombined09_T2starw.nii.gz: 44%|▍| 356k/805k [00:00<00:00, 1.42MB/
sub-MGH_rec-uncombined10_T2starw.nii.gz: 10%| | 85.2k/838k [00:00<00:01, 628kB/
sub-MGH_rec-uncombined11_T2starw.nii.gz: 10%| | 85.2k/857k [00:00<00:01, 607kB/
sub-MGH_rec-uncombined10_T2starw.nii.gz: 49%|▍| 408k/838k [00:00<00:00, 1.65MB/
sub-MGH_rec-uncombined12_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined11_T2starw.nii.gz: 48%|▍| 408k/857k [00:00<00:00, 1.55MB/
sub-MGH_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/861k [00:00<?, ?B/s]
sub-MGH_rec-uncombined13_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined12_T2starw.nii.gz: 10%| | 83.6k/861k [00:00<00:01, 633kB/
sub-MGH_rec-uncombined14_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined12_T2starw.nii.gz: 41%|▍| 356k/861k [00:00<00:00, 1.45MB/
sub-MGH_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/770k [00:00<?, ?B/s]
sub-MGH_rec-uncombined15_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/812k [00:00<?, ?B/s]
sub-MGH_rec-uncombined13_T2starw.nii.gz: 11%| | 83.5k/770k [00:00<00:01, 533kB/
sub-MGH_rec-uncombined16_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined14_T2starw.nii.gz: 11%| | 85.2k/812k [00:00<00:01, 613kB/
sub-MGH_rec-uncombined13_T2starw.nii.gz: 44%|▍| 339k/770k [00:00<00:00, 1.35MB/
sub-MGH_rec-uncombined14_T2starw.nii.gz: 44%|▍| 357k/812k [00:00<00:00, 1.51MB/
sub-MGH_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/743k [00:00<?, ?B/s]
sub-MGH_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/785k [00:00<?, ?B/s]
sub-MGH_rec-uncombined17_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined15_T2starw.nii.gz: 11%| | 85.2k/743k [00:00<00:01, 621kB/
sub-MGH_rec-uncombined16_T2starw.nii.gz: 11%| | 83.6k/785k [00:00<00:01, 622kB/
sub-MGH_rec-uncombined15_T2starw.nii.gz: 46%|▍| 340k/743k [00:00<00:00, 1.36MB/
sub-MGH_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/829k [00:00<?, ?B/s]
sub-MGH_rec-uncombined16_T2starw.nii.gz: 45%|▍| 356k/785k [00:00<00:00, 1.45MB/
sub-MGH_rec-uncombined18_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined17_T2starw.nii.gz: 8%| | 66.6k/829k [00:00<00:01, 672kB/
sub-MGH_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/739k [00:00<?, ?B/s]
sub-MGH_rec-uncombined17_T2starw.nii.gz: 20%|▏| 169k/829k [00:00<00:00, 845kB/s
sub-MGH_rec-uncombined18_T2starw.nii.gz: 11%| | 83.6k/739k [00:00<00:01, 616kB/
sub-MGH_rec-uncombined20_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined19_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH_rec-uncombined17_T2starw.nii.gz: 86%|▊| 713k/829k [00:00<00:00, 2.41MB/
sub-MGH_rec-uncombined18_T2starw.nii.gz: 37%|▎| 271k/739k [00:00<00:00, 1.20MB/
sub-MGH_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/684k [00:00<?, ?B/s]
sub-MGH_rec-uncombined19_T2starw.nii.gz: 12%| | 83.5k/684k [00:00<00:01, 608kB/
sub-MGH_acq-anat_TB1TFL.json: 0%| | 0.00/2.62k [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-MGH_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/795k [00:00<?, ?B/s]
sub-MGH_rec-uncombined19_T2starw.nii.gz: 59%|▌| 407k/684k [00:00<00:00, 1.62MB/
sub-MGH_rec-uncombined20_T2starw.nii.gz: 10%| | 77.0k/795k [00:00<00:01, 602kB/
sub-MGH_rec-uncombined20_T2starw.nii.gz: 46%|▍| 366k/795k [00:00<00:00, 1.52MB/
sub-MGH_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/939k [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.81M [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.60M [00:00<?, ?B/s]
sub-MGH_acq-anat_TB1TFL.nii.gz: 9%|▋ | 83.6k/939k [00:00<00:01, 602kB/s]
sub-MGH_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 85.2k/3.81M [00:00<00:06, 623kB/s
sub-MGH_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.60M [00:00<00:05, 619
sub-MGH_acq-anat_TB1TFL.nii.gz: 36%|██▉ | 339k/939k [00:00<00:00, 1.33MB/s]
sub-MGH_acq-coilQaSagLarge_SNR.nii.gz: 10%| | 374k/3.81M [00:00<00:02, 1.50MB/s
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.0M [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.60M [00:00<00:02, 1.43
sub-MGH_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH_acq-coilQaSagLarge_SNR.nii.gz: 35%|▎| 1.33M/3.81M [00:00<00:00, 4.47MB/
sub-MGH_acq-coilQaSagSmall_GFactor.nii.gz: 35%|▎| 1.26M/3.60M [00:00<00:00, 4.5
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/16.0M [00:00<00:27, 604kB/s]
sub-MGH_acq-coilQaSagLarge_SNR.nii.gz: 80%|▊| 3.04M/3.81M [00:00<00:00, 9.00MB/
sub-MGH_acq-coilQaSagSmall_GFactor.nii.gz: 74%|▋| 2.65M/3.60M [00:00<00:00, 8.0
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/16.0M [00:00<00:10, 1.62MB/s]
sub-MGH_acq-famp_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.33M/16.0M [00:00<00:03, 3.98MB/s
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 26%|▎| 4.12M/16.0M [00:00<00:01, 11.9MB/s
sub-MGH_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/85.7k [00:00<?, ?B/s]
sub-MGH_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/933k [00:00<?, ?B/s]
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 48%|▍| 7.69M/16.0M [00:00<00:00, 19.0MB/s
sub-MGH_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MGH_acq-famp-0.66_TB1DREAM.nii.gz: 98%|▉| 83.6k/85.7k [00:00<00:00, 605kB/s
sub-MGH_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/86.2k [00:00<?, ?B/s]
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 61%|▌| 9.69M/16.0M [00:00<00:00, 19.5MB/s
sub-MGH_acq-famp_TB1TFL.nii.gz: 9%|▋ | 83.6k/933k [00:00<00:01, 590kB/s]
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 84%|▊| 13.4M/16.0M [00:00<00:00, 25.1MB/s
sub-MGH_acq-famp_TB1DREAM.nii.gz: 99%|████▉| 85.2k/86.2k [00:00<00:00, 646kB/s]
sub-MGH_acq-famp_TB1TFL.nii.gz: 38%|███ | 356k/933k [00:00<00:00, 1.43MB/s]
sub-MGH_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MGH_acq-coilQaTra_GFactor.nii.gz: 100%|▉| 16.0M/16.0M [00:00<00:00, 24.6MB/s
sub-MGH_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.1k [00:00<?, ?B/s]
sub-MGH_acq-refv-0.66_TB1DREAM.nii.gz: 88%|▉| 77.0k/87.1k [00:00<00:00, 567kB/s
sub-MGH_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/91.4k [00:00<?, ?B/s]
sub-MNI_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH_acq-refv_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MNI_rec-uncombined01_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MGH_acq-refv_TB1DREAM.nii.gz: 93%|████▋| 85.2k/91.4k [00:00<00:00, 617kB/s]
sub-MNI_T2starw.nii.gz: 0%| | 0.00/1.14M [00:00<?, ?B/s]
sub-MNI_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/734k [00:00<?, ?B/s]
sub-MNI_rec-uncombined02_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined03_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_T2starw.nii.gz: 7%|█ | 83.5k/1.14M [00:00<00:01, 570kB/s]
sub-MNI_rec-uncombined01_T2starw.nii.gz: 7%| | 49.6k/734k [00:00<00:01, 447kB/
sub-MNI_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/742k [00:00<?, ?B/s]
sub-MNI_T2starw.nii.gz: 35%|█████▏ | 407k/1.14M [00:00<00:00, 1.63MB/s]
sub-MNI_rec-uncombined01_T2starw.nii.gz: 35%|▎| 254k/734k [00:00<00:00, 1.34MB/
sub-MNI_rec-uncombined02_T2starw.nii.gz: 11%| | 83.6k/742k [00:00<00:01, 614kB/
sub-MNI_rec-uncombined01_T2starw.nii.gz: 99%|▉| 730k/734k [00:00<00:00, 2.53MB/
sub-MNI_rec-uncombined04_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/835k [00:00<?, ?B/s]
sub-MNI_rec-uncombined02_T2starw.nii.gz: 48%|▍| 356k/742k [00:00<00:00, 1.43MB/
sub-MNI_rec-uncombined03_T2starw.nii.gz: 10%| | 83.6k/835k [00:00<00:01, 664kB/
sub-MNI_rec-uncombined05_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined03_T2starw.nii.gz: 43%|▍| 356k/835k [00:00<00:00, 1.46MB/
sub-MNI_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/792k [00:00<?, ?B/s]
sub-MNI_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/792k [00:00<?, ?B/s]
sub-MNI_rec-uncombined06_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined04_T2starw.nii.gz: 8%| | 66.5k/792k [00:00<00:01, 487kB/
sub-MNI_rec-uncombined05_T2starw.nii.gz: 11%| | 83.6k/792k [00:00<00:01, 675kB/
sub-MNI_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/801k [00:00<?, ?B/s]
sub-MNI_rec-uncombined04_T2starw.nii.gz: 34%|▎| 271k/792k [00:00<00:00, 1.06MB/
sub-MNI_rec-uncombined07_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined05_T2starw.nii.gz: 45%|▍| 356k/792k [00:00<00:00, 1.35MB/
sub-MNI_rec-uncombined06_T2starw.nii.gz: 10%| | 83.6k/801k [00:00<00:01, 574kB/
sub-MNI_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/817k [00:00<?, ?B/s]
sub-MNI_rec-uncombined06_T2starw.nii.gz: 42%|▍| 339k/801k [00:00<00:00, 1.37MB/
sub-MNI_rec-uncombined07_T2starw.nii.gz: 10%| | 83.6k/817k [00:00<00:01, 615kB/
sub-MNI_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/804k [00:00<?, ?B/s]
sub-MNI_rec-uncombined10_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined08_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined07_T2starw.nii.gz: 45%|▍| 368k/817k [00:00<00:00, 1.44MB/
sub-MNI_rec-uncombined08_T2starw.nii.gz: 10%| | 83.6k/804k [00:00<00:01, 617kB/
sub-MNI_rec-uncombined09_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined08_T2starw.nii.gz: 44%|▍| 356k/804k [00:00<00:00, 1.41MB/
sub-MNI_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/798k [00:00<?, ?B/s]
sub-MNI_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/816k [00:00<?, ?B/s]
sub-MNI_rec-uncombined11_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined09_T2starw.nii.gz: 10%| | 83.6k/798k [00:00<00:01, 659kB/
sub-MNI_rec-uncombined10_T2starw.nii.gz: 10%| | 83.6k/816k [00:00<00:01, 608kB/
sub-MNI_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/846k [00:00<?, ?B/s]
sub-MNI_rec-uncombined12_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined09_T2starw.nii.gz: 45%|▍| 356k/798k [00:00<00:00, 1.48MB/
sub-MNI_rec-uncombined10_T2starw.nii.gz: 44%|▍| 356k/816k [00:00<00:00, 1.41MB/
sub-MNI_rec-uncombined12_T2starw.nii.gz: 10%| | 83.6k/846k [00:00<00:01, 558kB/
sub-MNI_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/857k [00:00<?, ?B/s]
sub-MNI_rec-uncombined12_T2starw.nii.gz: 42%|▍| 356k/846k [00:00<00:00, 1.43MB/
sub-MNI_rec-uncombined13_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined11_T2starw.nii.gz: 10%| | 83.5k/857k [00:00<00:01, 564kB/
sub-MNI_rec-uncombined14_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/765k [00:00<?, ?B/s]
sub-MNI_rec-uncombined11_T2starw.nii.gz: 47%|▍| 407k/857k [00:00<00:00, 1.65MB/
sub-MNI_rec-uncombined13_T2starw.nii.gz: 11%| | 83.6k/765k [00:00<00:01, 663kB/
sub-MNI_rec-uncombined15_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/803k [00:00<?, ?B/s]
sub-MNI_rec-uncombined16_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined13_T2starw.nii.gz: 46%|▍| 356k/765k [00:00<00:00, 1.46MB/
sub-MNI_rec-uncombined14_T2starw.nii.gz: 11%| | 85.2k/803k [00:00<00:01, 704kB/
sub-MNI_rec-uncombined14_T2starw.nii.gz: 44%|▍| 357k/803k [00:00<00:00, 1.49MB/
sub-MNI_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/737k [00:00<?, ?B/s]
sub-MNI_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/728k [00:00<?, ?B/s]
sub-MNI_rec-uncombined17_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined15_T2starw.nii.gz: 11%| | 83.6k/737k [00:00<00:01, 566kB/
sub-MNI_rec-uncombined16_T2starw.nii.gz: 11%| | 83.6k/728k [00:00<00:01, 606kB/
sub-MNI_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/762k [00:00<?, ?B/s]
sub-MNI_rec-uncombined15_T2starw.nii.gz: 48%|▍| 356k/737k [00:00<00:00, 1.44MB/
sub-MNI_rec-uncombined16_T2starw.nii.gz: 47%|▍| 339k/728k [00:00<00:00, 1.34MB/
sub-MNI_rec-uncombined17_T2starw.nii.gz: 11%| | 83.6k/762k [00:00<00:01, 612kB/
sub-MNI_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/757k [00:00<?, ?B/s]
sub-MNI_rec-uncombined18_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined17_T2starw.nii.gz: 29%|▎| 220k/762k [00:00<00:00, 962kB/s
sub-MNI_rec-uncombined18_T2starw.nii.gz: 11%| | 83.6k/757k [00:00<00:01, 638kB/
sub-MNI_rec-uncombined18_T2starw.nii.gz: 47%|▍| 356k/757k [00:00<00:00, 1.45MB/
sub-MNI_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/635k [00:00<?, ?B/s]
sub-MNI_rec-uncombined20_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined19_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_rec-uncombined18_T2starw.nii.gz: 94%|▉| 713k/757k [00:00<00:00, 2.22MB/
sub-MNI_acq-anat_TB1TFL.json: 0%| | 0.00/2.59k [00:00<?, ?B/s]
sub-MNI_rec-uncombined19_T2starw.nii.gz: 12%| | 77.0k/635k [00:00<00:01, 565kB/
sub-MNI_rec-uncombined19_T2starw.nii.gz: 60%|▌| 383k/635k [00:00<00:00, 1.55MB/
sub-MNI_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-MNI_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/752k [00:00<?, ?B/s]
sub-MNI_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/942k [00:00<?, ?B/s]
sub-MNI_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.86M [00:00<?, ?B/s]
sub-MNI_rec-uncombined20_T2starw.nii.gz: 11%| | 83.5k/752k [00:00<00:01, 614kB/
sub-MNI_acq-anat_TB1TFL.nii.gz: 9%|▋ | 83.5k/942k [00:00<00:01, 610kB/s]
sub-MNI_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MNI_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.86M [00:00<00:07, 552kB/s
sub-MNI_acq-anat_TB1TFL.nii.gz: 29%|██▎ | 271k/942k [00:00<00:00, 1.27MB/s]
sub-MNI_rec-uncombined20_T2starw.nii.gz: 54%|▌| 407k/752k [00:00<00:00, 1.64MB/
sub-MNI_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.56M [00:00<?, ?B/s]
sub-MNI_acq-anat_TB1TFL.nii.gz: 88%|███████ | 832k/942k [00:00<00:00, 3.01MB/s]
sub-MNI_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 339k/3.86M [00:00<00:02, 1.28MB/s
sub-MNI_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 85.2k/3.56M [00:00<00:05, 682
sub-MNI_acq-coilQaSagLarge_SNR.nii.gz: 36%|▎| 1.39M/3.86M [00:00<00:00, 4.48MB/
sub-MNI_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MNI_acq-coilQaSagSmall_GFactor.nii.gz: 9%| | 340k/3.56M [00:00<00:02, 1.63
sub-MNI_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI_acq-coilQaSagSmall_GFactor.nii.gz: 20%|▏| 714k/3.56M [00:00<00:01, 2.45
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.9M [00:00<?, ?B/s]
sub-MNI_acq-coilQaSagSmall_GFactor.nii.gz: 62%|▌| 2.21M/3.56M [00:00<00:00, 7.1
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/15.9M [00:00<00:22, 725kB/s]
sub-MNI_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/15.9M [00:00<00:09, 1.75MB/s]
sub-MNI_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.2k [00:00<?, ?B/s]
sub-MNI_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/84.4k [00:00<?, ?B/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 9%| | 1.49M/15.9M [00:00<00:02, 5.54MB/s
sub-MNI_acq-famp_TB1DREAM.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-MNI_acq-famp-0.66_TB1DREAM.nii.gz: 95%|▉| 83.6k/88.2k [00:00<00:00, 549kB/s
sub-MNI_acq-famp-1.5_TB1DREAM.nii.gz: 80%|▊| 67.6k/84.4k [00:00<00:00, 680kB/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 13%|▏| 2.09M/15.9M [00:00<00:02, 5.58MB/s
sub-MNI_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/85.9k [00:00<?, ?B/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 39%|▍| 6.14M/15.9M [00:00<00:00, 17.7MB/s
sub-MNI_acq-famp_TB1DREAM.nii.gz: 97%|████▊| 83.5k/85.9k [00:00<00:00, 612kB/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 57%|▌| 9.09M/15.9M [00:00<00:00, 21.9MB/s
sub-MNI_acq-famp_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-MNI_acq-coilQaTra_GFactor.nii.gz: 84%|▊| 13.4M/15.9M [00:00<00:00, 28.1MB/s
sub-MNI_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/938k [00:00<?, ?B/s]
sub-MNI_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/85.1k [00:00<?, ?B/s]
sub-MNI_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MNI_acq-famp_TB1TFL.nii.gz: 9%|▋ | 83.6k/938k [00:00<00:01, 607kB/s]
sub-MNI_acq-refv-0.66_TB1DREAM.nii.gz: 98%|▉| 83.6k/85.1k [00:00<00:00, 604kB/s
sub-MNI_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MNI_acq-refv_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI_acq-famp_TB1TFL.nii.gz: 38%|███ | 356k/938k [00:00<00:00, 1.39MB/s]
sub-MNI_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.3k [00:00<?, ?B/s]
sub-MNI_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-MPI_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-MNI_acq-refv-1.5_TB1DREAM.nii.gz: 57%|▌| 52.2k/91.3k [00:00<00:00, 379kB/s]
sub-MPI_T2starw.nii.gz: 0%| | 0.00/1.16M [00:00<?, ?B/s]
sub-MNI_acq-refv_TB1DREAM.nii.gz: 93%|████▋| 83.6k/89.8k [00:00<00:00, 612kB/s]
sub-MPI_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/923k [00:00<?, ?B/s]
sub-MPI_T2starw.nii.gz: 7%|█ | 83.6k/1.16M [00:00<00:01, 610kB/s]
sub-MPI_rec-uncombined10_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/929k [00:00<?, ?B/s]
sub-MPI_T2starw.nii.gz: 30%|████▍ | 356k/1.16M [00:00<00:00, 1.42MB/s]
sub-MPI_rec-uncombined10_T2starw.nii.gz: 9%| | 83.6k/923k [00:00<00:01, 560kB/
sub-MPI_rec-uncombined11_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined11_T2starw.nii.gz: 9%| | 83.6k/929k [00:00<00:01, 616kB/
sub-MPI_rec-uncombined10_T2starw.nii.gz: 39%|▍| 356k/923k [00:00<00:00, 1.44MB/
sub-MPI_rec-uncombined12_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined11_T2starw.nii.gz: 38%|▍| 356k/929k [00:00<00:00, 1.35MB/
sub-MPI_rec-uncombined13_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/944k [00:00<?, ?B/s]
sub-MPI_rec-uncombined14_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined12_T2starw.nii.gz: 9%| | 85.2k/944k [00:00<00:01, 649kB/
sub-MPI_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/925k [00:00<?, ?B/s]
sub-MPI_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/966k [00:00<?, ?B/s]
sub-MPI_rec-uncombined12_T2starw.nii.gz: 38%|▍| 357k/944k [00:00<00:00, 1.50MB/
sub-MPI_rec-uncombined15_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined13_T2starw.nii.gz: 9%| | 83.6k/925k [00:00<00:01, 618kB/
sub-MPI_rec-uncombined12_T2starw.nii.gz: 99%|▉| 935k/944k [00:00<00:00, 3.00MB/
sub-MPI_rec-uncombined14_T2starw.nii.gz: 9%| | 85.2k/966k [00:00<00:01, 571kB/
sub-MPI_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/953k [00:00<?, ?B/s]
sub-MPI_rec-uncombined13_T2starw.nii.gz: 38%|▍| 356k/925k [00:00<00:00, 1.39MB/
sub-MPI_rec-uncombined14_T2starw.nii.gz: 42%|▍| 408k/966k [00:00<00:00, 1.66MB/
sub-MPI_rec-uncombined15_T2starw.nii.gz: 9%| | 85.2k/953k [00:00<00:01, 556kB/
sub-MPI_rec-uncombined16_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined17_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined15_T2starw.nii.gz: 43%|▍| 408k/953k [00:00<00:00, 1.67MB/
sub-MPI_rec-uncombined18_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/987k [00:00<?, ?B/s]
sub-MPI_rec-uncombined19_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 546kB
sub-MPI_rec-uncombined18_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 612kB
sub-MPI_rec-uncombined16_T2starw.nii.gz: 9%| | 85.2k/987k [00:00<00:01, 625kB/
sub-MPI_rec-uncombined18_T2starw.nii.gz: 17%|▏| 169k/0.99M [00:00<00:02, 321kB/
sub-MPI_rec-uncombined17_T2starw.nii.gz: 34%|▎| 340k/0.99M [00:00<00:01, 662kB/
sub-MPI_rec-uncombined16_T2starw.nii.gz: 17%|▏| 170k/987k [00:00<00:02, 344kB/s
sub-MPI_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI_rec-uncombined18_T2starw.nii.gz: 94%|▉| 952k/0.99M [00:00<00:00, 2.15MB
sub-MPI_rec-uncombined16_T2starw.nii.gz: 98%|▉| 969k/987k [00:00<00:00, 2.29MB/
sub-MPI_rec-uncombined19_T2starw.nii.gz: 18%|▏| 186k/1.01M [00:00<00:00, 1.26MB
sub-MPI_rec-uncombined1_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined19_T2starw.nii.gz: 72%|▋| 747k/1.01M [00:00<00:00, 3.04MB
sub-MPI_rec-uncombined21_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/990k [00:00<?, ?B/s]
sub-MPI_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/967k [00:00<?, ?B/s]
sub-MPI_rec-uncombined20_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined20_T2starw.nii.gz: 8%| | 83.5k/990k [00:00<00:01, 617kB/
sub-MPI_rec-uncombined1_T2starw.nii.gz: 9%| | 83.6k/967k [00:00<00:01, 614kB/s
sub-MPI_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/989k [00:00<?, ?B/s]
sub-MPI_rec-uncombined20_T2starw.nii.gz: 41%|▍| 407k/990k [00:00<00:00, 1.65MB/
sub-MPI_rec-uncombined1_T2starw.nii.gz: 40%|▍| 390k/967k [00:00<00:00, 1.57MB/s
sub-MPI_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/989k [00:00<00:01, 615kB/
sub-MPI_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-MPI_rec-uncombined23_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined21_T2starw.nii.gz: 34%|▎| 339k/989k [00:00<00:00, 1.36MB/
sub-MPI_rec-uncombined23_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 724kB
sub-MPI_rec-uncombined23_T2starw.nii.gz: 35%|▎| 356k/1.00M [00:00<00:00, 1.52MB
sub-MPI_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-MPI_rec-uncombined24_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined22_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined22_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 652kB
sub-MPI_rec-uncombined2_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined22_T2starw.nii.gz: 35%|▎| 356k/1.00M [00:00<00:00, 1.51MB
sub-MPI_rec-uncombined3_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/937k [00:00<?, ?B/s]
sub-MPI_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/983k [00:00<?, ?B/s]
sub-MPI_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/956k [00:00<?, ?B/s]
sub-MPI_rec-uncombined2_T2starw.nii.gz: 9%| | 85.2k/937k [00:00<00:01, 627kB/s
sub-MPI_rec-uncombined4_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined24_T2starw.nii.gz: 8%| | 83.6k/983k [00:00<00:01, 568kB/
sub-MPI_rec-uncombined3_T2starw.nii.gz: 9%| | 83.5k/956k [00:00<00:01, 612kB/s
sub-MPI_rec-uncombined5_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined2_T2starw.nii.gz: 38%|▍| 357k/937k [00:00<00:00, 1.43MB/s
sub-MPI_rec-uncombined24_T2starw.nii.gz: 36%|▎| 356k/983k [00:00<00:00, 1.43MB/
sub-MPI_rec-uncombined3_T2starw.nii.gz: 43%|▍| 407k/956k [00:00<00:00, 1.56MB/s
sub-MPI_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/964k [00:00<?, ?B/s]
sub-MPI_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/960k [00:00<?, ?B/s]
sub-MPI_rec-uncombined5_T2starw.nii.gz: 9%| | 83.5k/964k [00:00<00:01, 612kB/s
sub-MPI_rec-uncombined6_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined8_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/930k [00:00<?, ?B/s]
sub-MPI_rec-uncombined5_T2starw.nii.gz: 33%|▎| 322k/964k [00:00<00:00, 1.53MB/s
sub-MPI_rec-uncombined4_T2starw.nii.gz: 9%| | 85.2k/960k [00:00<00:01, 625kB/s
sub-MPI_rec-uncombined5_T2starw.nii.gz: 86%|▊| 832k/964k [00:00<00:00, 2.95MB/s
sub-MPI_rec-uncombined6_T2starw.nii.gz: 9%| | 83.5k/930k [00:00<00:01, 599kB/s
sub-MPI_rec-uncombined4_T2starw.nii.gz: 37%|▎| 357k/960k [00:00<00:00, 1.43MB/s
sub-MPI_rec-uncombined7_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined6_T2starw.nii.gz: 44%|▍| 407k/930k [00:00<00:00, 1.56MB/s
sub-MPI_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-MPI_rec-uncombined7_T2starw.nii.gz: 9%| | 83.6k/968k [00:00<00:01, 614kB/s
sub-MPI_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/951k [00:00<?, ?B/s]
sub-MPI_rec-uncombined9_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MPI_rec-uncombined7_T2starw.nii.gz: 35%|▎| 339k/968k [00:00<00:00, 1.35MB/s
sub-MPI_rec-uncombined8_T2starw.nii.gz: 9%| | 83.6k/951k [00:00<00:01, 610kB/s
sub-MPI_rec-uncombined9_T2starw.nii.gz: 0%| | 0.00/925k [00:00<?, ?B/s]
sub-MPI_rec-uncombined8_T2starw.nii.gz: 32%|▎| 306k/951k [00:00<00:00, 1.42MB/s
sub-MPI_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/702k [00:00<?, ?B/s]
sub-MPI_rec-uncombined9_T2starw.nii.gz: 9%| | 83.6k/925k [00:00<00:01, 615kB/s
sub-MPI_rec-uncombined8_T2starw.nii.gz: 79%|▊| 747k/951k [00:00<00:00, 2.60MB/s
sub-MPI_rec-uncombined9_T2starw.nii.gz: 29%|▎| 271k/925k [00:00<00:00, 1.24MB/s
sub-MPI_acq-anat_TB1TFL.nii.gz: 12%|▉ | 83.6k/702k [00:00<00:01, 605kB/s]
sub-MPI_acq-anat_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-MPI_rec-uncombined9_T2starw.nii.gz: 79%|▊| 730k/925k [00:00<00:00, 2.61MB/s
sub-MPI_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MPI_acq-anat_TB1TFL.nii.gz: 51%|████ | 356k/702k [00:00<00:00, 1.44MB/s]
sub-MPI_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.99M [00:00<?, ?B/s]
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.5M [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.99M [00:00<00:05, 728kB/s
sub-MPI_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.53M [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 7%| | 289k/3.99M [00:00<00:02, 1.42MB/s
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/15.5M [00:00<00:26, 617kB/s]
sub-MPI_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.4k [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 18%|▏| 731k/3.99M [00:00<00:01, 2.71MB/s
sub-MPI_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.53M [00:00<00:05, 690
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/15.5M [00:00<00:11, 1.43MB/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 63%|▋| 2.51M/3.99M [00:00<00:00, 8.79MB/
sub-MPI_acq-famp-0.66_TB1DREAM.nii.gz: 96%|▉| 83.5k/87.4k [00:00<00:00, 610kB/s
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 5%| | 849k/15.5M [00:00<00:05, 2.79MB/s]
sub-MPI_acq-coilQaSagSmall_GFactor.nii.gz: 9%| | 340k/3.53M [00:00<00:02, 1.39
sub-MPI_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.9k [00:00<?, ?B/s]
sub-MPI_acq-coilQaSagLarge_SNR.nii.gz: 94%|▉| 3.76M/3.99M [00:00<00:00, 10.3MB/
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 12%| | 1.89M/15.5M [00:00<00:02, 5.67MB/s
sub-MPI_acq-coilQaSagSmall_GFactor.nii.gz: 30%|▎| 1.06M/3.53M [00:00<00:00, 3.7
sub-MPI_acq-famp-1.5_TB1DREAM.nii.gz: 95%|▉| 85.2k/89.9k [00:00<00:00, 624kB/s]
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 27%|▎| 4.12M/15.5M [00:00<00:01, 11.6MB/s
sub-MPI_acq-coilQaSagSmall_GFactor.nii.gz: 69%|▋| 2.45M/3.53M [00:00<00:00, 7.5
sub-MPI_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 37%|▎| 5.71M/15.5M [00:00<00:00, 13.2MB/s
sub-MPI_acq-famp_TB1DREAM.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 64%|▋| 9.85M/15.5M [00:00<00:00, 22.7MB/s
sub-MPI_acq-coilQaTra_GFactor.nii.gz: 96%|▉| 14.8M/15.5M [00:00<00:00, 31.6MB/s
sub-MPI_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MPI_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/87.5k [00:00<?, ?B/s]
sub-MPI_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/933k [00:00<?, ?B/s]
sub-MPI_acq-famp_TB1DREAM.nii.gz: 95%|████▊| 83.5k/87.5k [00:00<00:00, 696kB/s]
sub-MPI_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/80.7k [00:00<?, ?B/s]
sub-MPI_acq-famp_TB1TFL.nii.gz: 8%|▋ | 77.0k/933k [00:00<00:01, 568kB/s]
sub-MPI_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-MPI_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 80.7k/80.7k [00:00<00:00, 622kB/s
sub-MPI_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-MPI_acq-famp_TB1TFL.nii.gz: 37%|██▉ | 349k/933k [00:00<00:00, 1.38MB/s]
sub-MPI_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/93.2k [00:00<?, ?B/s]
sub-MSSM_T2starw.json: 0%| | 0.00/2.53k [00:00<?, ?B/s]
sub-MPI_acq-refv_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/84.7k [00:00<?, ?B/s]
sub-MPI_acq-refv-1.5_TB1DREAM.nii.gz: 90%|▉| 83.6k/93.2k [00:00<00:00, 609kB/s]
sub-MSSM_T2starw.nii.gz: 0%| | 0.00/1.74M [00:00<?, ?B/s]
sub-MPI_acq-refv_TB1DREAM.nii.gz: 100%|█████| 84.7k/84.7k [00:00<00:00, 664kB/s]
sub-MSSM_rec-uncombined10_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_T2starw.nii.gz: 5%|▋ | 83.5k/1.74M [00:00<00:02, 604kB/s]
sub-MSSM_rec-uncombined11_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/974k [00:00<?, ?B/s]
sub-MSSM_T2starw.nii.gz: 23%|███▏ | 407k/1.74M [00:00<00:00, 1.63MB/s]
sub-MSSM_rec-uncombined10_T2starw.nii.gz: 9%| | 83.6k/974k [00:00<00:01, 605kB
sub-MSSM_rec-uncombined12_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/934k [00:00<?, ?B/s]
sub-MSSM_T2starw.nii.gz: 66%|████████▌ | 1.14M/1.74M [00:00<00:00, 3.79MB/s]
sub-MSSM_rec-uncombined10_T2starw.nii.gz: 26%|▎| 254k/974k [00:00<00:00, 1.17MB
sub-MSSM_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/826k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined11_T2starw.nii.gz: 9%| | 83.6k/934k [00:00<00:01, 560kB
sub-MSSM_rec-uncombined10_T2starw.nii.gz: 77%|▊| 747k/974k [00:00<00:00, 2.65MB
sub-MSSM_rec-uncombined12_T2starw.nii.gz: 10%| | 83.6k/826k [00:00<00:01, 607kB
sub-MSSM_rec-uncombined11_T2starw.nii.gz: 38%|▍| 356k/934k [00:00<00:00, 1.46MB
sub-MSSM_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/1.12M [00:00<?, ?B/s]
sub-MSSM_rec-uncombined13_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined11_T2starw.nii.gz: 85%|▊| 798k/934k [00:00<00:00, 2.54MB
sub-MSSM_rec-uncombined12_T2starw.nii.gz: 43%|▍| 356k/826k [00:00<00:00, 1.39MB
sub-MSSM_rec-uncombined13_T2starw.nii.gz: 7%| | 77.0k/1.12M [00:00<00:01, 566k
sub-MSSM_rec-uncombined14_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined13_T2starw.nii.gz: 32%|▎| 366k/1.12M [00:00<00:00, 1.47M
sub-MSSM_rec-uncombined15_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/949k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined16_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined14_T2starw.nii.gz: 9%| | 85.2k/949k [00:00<00:01, 691kB
sub-MSSM_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/934k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined14_T2starw.nii.gz: 38%|▍| 357k/949k [00:00<00:00, 1.49MB
sub-MSSM_rec-uncombined17_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined15_T2starw.nii.gz: 9%| | 83.6k/934k [00:00<00:01, 510kB
sub-MSSM_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-MSSM_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/721k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined15_T2starw.nii.gz: 38%|▍| 356k/934k [00:00<00:00, 1.45MB
sub-MSSM_rec-uncombined16_T2starw.nii.gz: 7%| | 77.0k/1.06M [00:00<00:01, 565k
sub-MSSM_rec-uncombined17_T2starw.nii.gz: 12%| | 83.6k/721k [00:00<00:01, 606kB
sub-MSSM_rec-uncombined18_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined16_T2starw.nii.gz: 34%|▎| 366k/1.06M [00:00<00:00, 1.44M
sub-MSSM_rec-uncombined17_T2starw.nii.gz: 47%|▍| 340k/721k [00:00<00:00, 1.62MB
sub-MSSM_rec-uncombined19_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/695k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined1_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined18_T2starw.nii.gz: 12%| | 85.2k/695k [00:00<00:00, 647kB
sub-MSSM_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/927k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined20_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined18_T2starw.nii.gz: 51%|▌| 357k/695k [00:00<00:00, 1.52MB
sub-MSSM_rec-uncombined1_T2starw.nii.gz: 9%| | 83.6k/927k [00:00<00:01, 610kB/
sub-MSSM_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/896k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined1_T2starw.nii.gz: 38%|▍| 356k/927k [00:00<00:00, 1.42MB/
sub-MSSM_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/582k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined21_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined19_T2starw.nii.gz: 9%| | 77.0k/896k [00:00<00:01, 564kB
sub-MSSM_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/831k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined20_T2starw.nii.gz: 14%|▏| 83.6k/582k [00:00<00:00, 607kB
sub-MSSM_rec-uncombined19_T2starw.nii.gz: 39%|▍| 349k/896k [00:00<00:00, 1.40MB
sub-MSSM_rec-uncombined21_T2starw.nii.gz: 10%| | 85.2k/831k [00:00<00:01, 621kB
sub-MSSM_rec-uncombined20_T2starw.nii.gz: 61%|▌| 356k/582k [00:00<00:00, 1.41MB
sub-MSSM_rec-uncombined22_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined19_T2starw.nii.gz: 88%|▉| 791k/896k [00:00<00:00, 2.58MB
sub-MSSM_rec-uncombined2_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined21_T2starw.nii.gz: 33%|▎| 272k/831k [00:00<00:00, 1.22MB
sub-MSSM_rec-uncombined21_T2starw.nii.gz: 78%|▊| 646k/831k [00:00<00:00, 2.26MB
sub-MSSM_rec-uncombined3_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/833k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/901k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/871k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/891k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined22_T2starw.nii.gz: 10%| | 83.6k/833k [00:00<00:01, 615kB
sub-MSSM_rec-uncombined2_T2starw.nii.gz: 10%| | 83.6k/871k [00:00<00:01, 613kB/
sub-MSSM_rec-uncombined3_T2starw.nii.gz: 9%| | 83.6k/901k [00:00<00:01, 608kB/
sub-MSSM_rec-uncombined4_T2starw.nii.gz: 10%| | 85.2k/891k [00:00<00:01, 621kB/
sub-MSSM_rec-uncombined4_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined2_T2starw.nii.gz: 43%|▍| 373k/871k [00:00<00:00, 1.53MB/
sub-MSSM_rec-uncombined22_T2starw.nii.gz: 41%|▍| 339k/833k [00:00<00:00, 1.35MB
sub-MSSM_rec-uncombined3_T2starw.nii.gz: 39%|▍| 356k/901k [00:00<00:00, 1.42MB/
sub-MSSM_rec-uncombined2_T2starw.nii.gz: 100%|█| 871k/871k [00:00<00:00, 2.81MB/
sub-MSSM_rec-uncombined3_T2starw.nii.gz: 90%|▉| 815k/901k [00:00<00:00, 2.62MB/
sub-MSSM_rec-uncombined4_T2starw.nii.gz: 40%|▍| 357k/891k [00:00<00:00, 1.37MB/
sub-MSSM_rec-uncombined5_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined6_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/980k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-MSSM_rec-uncombined7_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined5_T2starw.nii.gz: 9%| | 83.6k/980k [00:00<00:01, 612kB/
sub-MSSM_rec-uncombined6_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 611kB
sub-MSSM_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/756k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined5_T2starw.nii.gz: 40%|▍| 390k/980k [00:00<00:00, 1.57MB/
sub-MSSM_rec-uncombined6_T2starw.nii.gz: 39%|▍| 390k/0.98M [00:00<00:00, 1.46MB
sub-MSSM_rec-uncombined8_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined7_T2starw.nii.gz: 9%| | 68.2k/756k [00:00<00:01, 645kB/
sub-MSSM_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/858k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined7_T2starw.nii.gz: 23%|▏| 170k/756k [00:00<00:00, 868kB/s
sub-MSSM_rec-uncombined8_T2starw.nii.gz: 10%| | 83.6k/858k [00:00<00:01, 606kB/
sub-MSSM_rec-uncombined7_T2starw.nii.gz: 94%|▉| 714k/756k [00:00<00:00, 2.51MB/
sub-MSSM_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined8_T2starw.nii.gz: 41%|▍| 356k/858k [00:00<00:00, 1.28MB/
sub-MSSM_rec-uncombined9_T2starw.nii.gz: 0%| | 0.00/949k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined9_T2starw.json: 0%| | 0.00/2.36k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined9_T2starw.nii.gz: 9%| | 83.6k/949k [00:00<00:01, 721kB/
sub-MSSM_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MSSM_rec-uncombined9_T2starw.nii.gz: 41%|▍| 390k/949k [00:00<00:00, 1.53MB/
sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.94M [00:00<?, ?B/s]
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.8M [00:00<?, ?B/s]
sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 85.2k/3.94M [00:00<00:06, 661kB/
sub-MSSM_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/4.19M [00:00<?, ?B/s]
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 0%| | 83.5k/16.8M [00:00<00:28, 612kB/s
sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 340k/3.94M [00:00<00:02, 1.38MB/
sub-MSSM_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/4.19M [00:00<00:07, 60
sub-MSSM_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz: 30%|▎| 1.20M/3.94M [00:00<00:00, 4.09MB
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/16.8M [00:00<00:10, 1.64MB/s
sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz: 73%|▋| 2.86M/3.94M [00:00<00:00, 8.61MB
sub-MSSM_acq-coilQaSagSmall_GFactor.nii.gz: 8%| | 356k/4.19M [00:00<00:02, 1.4
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.28M/16.8M [00:00<00:03, 4.48MB/
sub-MSSM_acq-coilQaSagSmall_GFactor.nii.gz: 26%|▎| 1.09M/4.19M [00:00<00:00, 3.
sub-MSSM_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/89.1k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 15%|▏| 2.57M/16.8M [00:00<00:01, 7.64MB/
sub-MSSM_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaSagSmall_GFactor.nii.gz: 56%|▌| 2.33M/4.19M [00:00<00:00, 6.
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 28%|▎| 4.65M/16.8M [00:00<00:01, 12.3MB/
sub-MSSM_acq-famp-0.66_TB1DREAM.nii.gz: 94%|▉| 83.6k/89.1k [00:00<00:00, 615kB/
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 37%|▎| 6.18M/16.8M [00:00<00:00, 13.2MB/
sub-MSSM_acq-famp_TB1DREAM.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 60%|▌| 9.99M/16.8M [00:00<00:00, 21.5MB/
sub-MSSM_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.9k [00:00<?, ?B/s]
sub-MSSM_acq-coilQaTra_GFactor.nii.gz: 84%|▊| 14.0M/16.8M [00:00<00:00, 27.2MB/
sub-MSSM_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-MSSM_acq-famp_TB1TFL.json: 0%| | 0.00/2.50k [00:00<?, ?B/s]
sub-MSSM_acq-famp-1.5_TB1DREAM.nii.gz: 58%|▌| 52.2k/89.9k [00:00<00:00, 490kB/s
sub-MSSM_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/809k [00:00<?, ?B/s]
sub-MSSM_acq-famp_TB1DREAM.nii.gz: 95%|███▊| 85.2k/89.8k [00:00<00:00, 628kB/s]
sub-MSSM_acq-famp_TB1TFL.nii.gz: 10%|▋ | 83.6k/809k [00:00<00:01, 611kB/s]
sub-MSSM_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MSSM_acq-famp_TB1TFL.nii.gz: 40%|██▊ | 322k/809k [00:00<00:00, 1.27MB/s]
sub-MSSM_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MSSM_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.2k [00:00<?, ?B/s]
sub-MSSM_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/90.7k [00:00<?, ?B/s]
sub-MSSM_acq-refv-0.66_TB1DREAM.nii.gz: 88%|▉| 76.5k/87.2k [00:00<00:00, 599kB/
sub-MSSM_acq-refv-1.5_TB1DREAM.nii.gz: 92%|▉| 83.6k/90.7k [00:00<00:00, 603kB/s
sub-MSSM_acq-refv_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MSSM_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-NTNU_T2starw.json: 0%| | 0.00/2.47k [00:00<?, ?B/s]
sub-NTNU_T2starw.nii.gz: 0%| | 0.00/1.18M [00:00<?, ?B/s]
sub-MSSM_acq-refv_TB1DREAM.nii.gz: 93%|███▋| 83.6k/89.8k [00:00<00:00, 554kB/s]
sub-NTNU_rec-uncombined01_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_T2starw.nii.gz: 6%|▉ | 78.0k/1.18M [00:00<00:01, 602kB/s]
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/981k [00:00<?, ?B/s]
sub-NTNU_T2starw.nii.gz: 29%|████ | 349k/1.18M [00:00<00:00, 1.44MB/s]
sub-NTNU_rec-uncombined02_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 7%| | 67.6k/981k [00:00<00:02, 415kB
sub-NTNU_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/939k [00:00<?, ?B/s]
sub-NTNU_T2starw.nii.gz: 94%|████████████▏| 1.10M/1.18M [00:00<00:00, 3.42MB/s]
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 17%|▏| 169k/981k [00:00<00:01, 630kB/
sub-NTNU_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/988k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined02_T2starw.nii.gz: 9%| | 85.2k/939k [00:00<00:01, 619kB
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 28%|▎| 271k/981k [00:00<00:01, 682kB/
sub-NTNU_rec-uncombined03_T2starw.nii.gz: 8%| | 83.6k/988k [00:00<00:01, 605kB
sub-NTNU_rec-uncombined03_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined02_T2starw.nii.gz: 38%|▍| 357k/939k [00:00<00:00, 1.41MB
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 38%|▍| 373k/981k [00:00<00:00, 714kB/
sub-NTNU_rec-uncombined04_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined03_T2starw.nii.gz: 36%|▎| 356k/988k [00:00<00:00, 1.36MB
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 52%|▌| 509k/981k [00:00<00:00, 808kB/
sub-NTNU_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/975k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 66%|▋| 645k/981k [00:00<00:00, 870kB/
sub-NTNU_rec-uncombined06_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined04_T2starw.nii.gz: 9%| | 83.6k/975k [00:00<00:01, 610kB
sub-NTNU_rec-uncombined05_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 81%|▊| 798k/981k [00:01<00:00, 928kB/
sub-NTNU_rec-uncombined05_T2starw.nii.gz: 9%| | 85.2k/968k [00:00<00:01, 623kB
sub-NTNU_rec-uncombined04_T2starw.nii.gz: 36%|▎| 356k/975k [00:00<00:00, 1.41MB
sub-NTNU_rec-uncombined01_T2starw.nii.gz: 97%|▉| 951k/981k [00:01<00:00, 1.01MB
sub-NTNU_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/961k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined05_T2starw.nii.gz: 37%|▎| 357k/968k [00:00<00:00, 1.42MB
sub-NTNU_rec-uncombined07_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined06_T2starw.nii.gz: 10%| | 94.0k/961k [00:00<00:01, 784kB
sub-NTNU_rec-uncombined06_T2starw.nii.gz: 42%|▍| 400k/961k [00:00<00:00, 1.67MB
sub-NTNU_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/944k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined08_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined06_T2starw.nii.gz: 100%|█| 961k/961k [00:00<00:00, 3.24MB
sub-NTNU_rec-uncombined09_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined07_T2starw.nii.gz: 6%| | 61.0k/0.99M [00:00<00:01, 587k
sub-NTNU_rec-uncombined08_T2starw.nii.gz: 8%| | 77.0k/944k [00:00<00:01, 562kB
sub-NTNU_rec-uncombined07_T2starw.nii.gz: 16%|▏| 162k/0.99M [00:00<00:01, 824kB
sub-NTNU_rec-uncombined08_T2starw.nii.gz: 37%|▎| 349k/944k [00:00<00:00, 1.39MB
sub-NTNU_rec-uncombined07_T2starw.nii.gz: 71%|▋| 723k/0.99M [00:00<00:00, 2.75M
sub-NTNU_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined10_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined08_T2starw.nii.gz: 80%|▊| 757k/944k [00:00<00:00, 2.27MB
sub-NTNU_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/998k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined09_T2starw.nii.gz: 8%| | 77.0k/0.99M [00:00<00:01, 565k
sub-NTNU_rec-uncombined10_T2starw.nii.gz: 8%| | 83.6k/998k [00:00<00:01, 608kB
sub-NTNU_rec-uncombined11_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined09_T2starw.nii.gz: 38%|▍| 383k/0.99M [00:00<00:00, 1.42M
sub-NTNU_rec-uncombined10_T2starw.nii.gz: 29%|▎| 288k/998k [00:00<00:00, 1.35MB
sub-NTNU_rec-uncombined12_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined10_T2starw.nii.gz: 73%|▋| 730k/998k [00:00<00:00, 2.53MB
sub-NTNU_rec-uncombined11_T2starw.nii.gz: 8%| | 85.2k/1.01M [00:00<00:01, 774k
sub-NTNU_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined13_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined11_T2starw.nii.gz: 35%|▎| 357k/1.01M [00:00<00:00, 1.55M
sub-NTNU_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/999k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined12_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 559k
sub-NTNU_rec-uncombined14_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined13_T2starw.nii.gz: 8%| | 83.6k/999k [00:00<00:01, 613kB
sub-NTNU_rec-uncombined12_T2starw.nii.gz: 35%|▎| 356k/1.00M [00:00<00:00, 1.44M
sub-NTNU_rec-uncombined13_T2starw.nii.gz: 36%|▎| 356k/999k [00:00<00:00, 1.42MB
sub-NTNU_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined15_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined16_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined14_T2starw.nii.gz: 8%| | 85.2k/1.03M [00:00<00:01, 625k
sub-NTNU_rec-uncombined14_T2starw.nii.gz: 34%|▎| 357k/1.03M [00:00<00:00, 1.42M
sub-NTNU_rec-uncombined17_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/998k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/932k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined15_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 713k
sub-NTNU_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/998k [00:00<00:01, 637kB
sub-NTNU_rec-uncombined17_T2starw.nii.gz: 9%| | 85.2k/932k [00:00<00:01, 622kB
sub-NTNU_rec-uncombined15_T2starw.nii.gz: 35%|▎| 356k/0.99M [00:00<00:00, 1.50M
sub-NTNU_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/919k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined16_T2starw.nii.gz: 39%|▍| 390k/998k [00:00<00:00, 1.57MB
sub-NTNU_rec-uncombined18_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined17_T2starw.nii.gz: 38%|▍| 357k/932k [00:00<00:00, 1.29MB
sub-NTNU_rec-uncombined18_T2starw.nii.gz: 9%| | 83.6k/919k [00:00<00:01, 544kB
sub-NTNU_rec-uncombined18_T2starw.nii.gz: 39%|▍| 356k/919k [00:00<00:00, 1.44MB
sub-NTNU_rec-uncombined19_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined20_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/971k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/947k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined19_T2starw.nii.gz: 9%| | 83.6k/971k [00:00<00:01, 644kB
sub-NTNU_rec-uncombined20_T2starw.nii.gz: 9%| | 83.6k/947k [00:00<00:01, 609kB
sub-NTNU_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/949k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined21_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined22_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined19_T2starw.nii.gz: 37%|▎| 356k/971k [00:00<00:00, 1.40MB
sub-NTNU_rec-uncombined20_T2starw.nii.gz: 34%|▎| 322k/947k [00:00<00:00, 1.53MB
sub-NTNU_rec-uncombined21_T2starw.nii.gz: 8%| | 77.0k/949k [00:00<00:01, 518kB
sub-NTNU_rec-uncombined20_T2starw.nii.gz: 77%|▊| 730k/947k [00:00<00:00, 2.51MB
sub-NTNU_rec-uncombined21_T2starw.nii.gz: 39%|▍| 366k/949k [00:00<00:00, 1.45MB
sub-NTNU_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/948k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined23_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/944k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined24_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined22_T2starw.nii.gz: 9%| | 83.5k/948k [00:00<00:01, 685kB
sub-NTNU_rec-uncombined23_T2starw.nii.gz: 9%| | 83.6k/944k [00:00<00:01, 668kB
sub-NTNU_rec-uncombined22_T2starw.nii.gz: 43%|▍| 407k/948k [00:00<00:00, 1.71MB
sub-NTNU_rec-uncombined23_T2starw.nii.gz: 38%|▍| 356k/944k [00:00<00:00, 1.47MB
sub-NTNU_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/951k [00:00<?, ?B/s]
sub-NTNU_acq-anat_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-NTNU_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/727k [00:00<?, ?B/s]
sub-NTNU_rec-uncombined24_T2starw.nii.gz: 8%| | 77.0k/951k [00:00<00:01, 570kB
sub-NTNU_acq-anat_TB1TFL.nii.gz: 11%|▊ | 83.6k/727k [00:00<00:01, 607kB/s]
sub-NTNU_rec-uncombined24_T2starw.nii.gz: 38%|▍| 366k/951k [00:00<00:00, 1.48MB
sub-NTNU_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.23k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-NTNU_acq-anat_TB1TFL.nii.gz: 49%|███▍ | 356k/727k [00:00<00:00, 1.32MB/s]
sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.93M [00:00<?, ?B/s]
sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.93M [00:00<00:06, 611kB/
sub-NTNU_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 356k/3.93M [00:00<00:02, 1.39MB/
sub-NTNU_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.58M [00:00<?, ?B/s]
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.5M [00:00<?, ?B/s]
sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz: 37%|▎| 1.46M/3.93M [00:00<00:00, 4.58MB
sub-NTNU_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.58M [00:00<00:05, 61
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/15.5M [00:00<00:26, 605kB/s
sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz: 99%|▉| 3.90M/3.93M [00:00<00:00, 11.3MB
sub-NTNU_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.3k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.58M [00:00<00:02, 1.4
sub-NTNU_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 3%| | 407k/15.5M [00:00<00:10, 1.58MB/s
sub-NTNU_acq-famp-0.66_TB1DREAM.nii.gz: 88%|▉| 77.0k/87.3k [00:00<00:00, 570kB/
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 6%| | 986k/15.5M [00:00<00:04, 3.17MB/s
sub-NTNU_acq-coilQaSagSmall_GFactor.nii.gz: 41%|▍| 1.46M/3.58M [00:00<00:00, 4.
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 18%|▏| 2.77M/15.5M [00:00<00:01, 7.90MB/
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 49%|▍| 7.60M/15.5M [00:00<00:00, 21.2MB/
sub-NTNU_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/88.2k [00:00<?, ?B/s]
sub-NTNU_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-NTNU_acq-famp_TB1DREAM.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 68%|▋| 10.5M/15.5M [00:00<00:00, 23.9MB/
sub-NTNU_acq-famp_TB1TFL.json: 0%| | 0.00/2.73k [00:00<?, ?B/s]
sub-NTNU_acq-famp_TB1DREAM.nii.gz: 87%|███▍| 77.0k/88.2k [00:00<00:00, 568kB/s]
sub-NTNU_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 83.6k/89.8k [00:00<00:00, 580kB/s
sub-NTNU_acq-coilQaTra_GFactor.nii.gz: 85%|▊| 13.1M/15.5M [00:00<00:00, 24.5MB/
sub-NTNU_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-NTNU_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/81.5k [00:00<?, ?B/s]
sub-NTNU_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-NTNU_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/910k [00:00<?, ?B/s]
sub-NTNU_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/93.4k [00:00<?, ?B/s]
sub-NTNU_acq-refv_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU_acq-refv-0.66_TB1DREAM.nii.gz: 94%|▉| 77.0k/81.5k [00:00<00:00, 564kB/
sub-NTNU_acq-famp_TB1TFL.nii.gz: 8%|▌ | 77.0k/910k [00:00<00:01, 582kB/s]
sub-NTNU_acq-refv-1.5_TB1DREAM.nii.gz: 89%|▉| 83.6k/93.4k [00:00<00:00, 618kB/s
sub-NTNU_acq-famp_TB1TFL.nii.gz: 35%|██▍ | 315k/910k [00:00<00:00, 1.53MB/s]
sub-NTNU_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/86.1k [00:00<?, ?B/s]
sub-NTNU_acq-famp_TB1TFL.nii.gz: 81%|█████▋ | 740k/910k [00:00<00:00, 2.70MB/s]
sub-UCL_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-UCL_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-NTNU_acq-refv_TB1DREAM.nii.gz: 97%|███▉| 83.6k/86.1k [00:00<00:00, 612kB/s]
sub-UCL_T2starw.nii.gz: 8%|█▏ | 85.2k/1.01M [00:00<00:01, 622kB/s]
sub-UCL_rec-uncombined1_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_T2starw.nii.gz: 35%|█████▏ | 357k/1.01M [00:00<00:00, 1.42MB/s]
sub-UCL_rec-uncombined2_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/860k [00:00<?, ?B/s]
sub-UCL_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/841k [00:00<?, ?B/s]
sub-UCL_rec-uncombined1_T2starw.nii.gz: 10%| | 83.5k/860k [00:00<00:01, 629kB/s
sub-UCL_rec-uncombined3_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined2_T2starw.nii.gz: 10%| | 83.6k/841k [00:00<00:01, 643kB/s
sub-UCL_rec-uncombined1_T2starw.nii.gz: 43%|▍| 373k/860k [00:00<00:00, 1.51MB/s
sub-UCL_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/813k [00:00<?, ?B/s]
sub-UCL_rec-uncombined2_T2starw.nii.gz: 42%|▍| 356k/841k [00:00<00:00, 1.46MB/s
sub-UCL_rec-uncombined4_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined1_T2starw.nii.gz: 100%|█| 860k/860k [00:00<00:00, 2.80MB/s
sub-UCL_rec-uncombined3_T2starw.nii.gz: 10%| | 83.5k/813k [00:00<00:01, 619kB/s
sub-UCL_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/796k [00:00<?, ?B/s]
sub-UCL_rec-uncombined3_T2starw.nii.gz: 42%|▍| 339k/813k [00:00<00:00, 1.36MB/s
sub-UCL_rec-uncombined4_T2starw.nii.gz: 11%| | 85.2k/796k [00:00<00:01, 627kB/s
sub-UCL_rec-uncombined5_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined6_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined3_T2starw.nii.gz: 92%|▉| 747k/813k [00:00<00:00, 2.27MB/s
sub-UCL_rec-uncombined4_T2starw.nii.gz: 32%|▎| 255k/796k [00:00<00:00, 1.09MB/s
sub-UCL_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/935k [00:00<?, ?B/s]
sub-UCL_rec-uncombined5_T2starw.nii.gz: 9%| | 83.6k/935k [00:00<00:01, 719kB/s
sub-UCL_rec-uncombined7_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined5_T2starw.nii.gz: 38%|▍| 356k/935k [00:00<00:00, 1.50MB/s
sub-UCL_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/821k [00:00<?, ?B/s]
sub-UCL_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/788k [00:00<?, ?B/s]
sub-UCL_rec-uncombined8_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-UCL_rec-uncombined6_T2starw.nii.gz: 10%| | 83.5k/821k [00:00<00:01, 653kB/s
sub-UCL_rec-uncombined7_T2starw.nii.gz: 11%| | 85.2k/788k [00:00<00:01, 625kB/s
sub-UCL_rec-uncombined6_T2starw.nii.gz: 41%|▍| 340k/821k [00:00<00:00, 1.68MB/s
sub-UCL_rec-uncombined7_T2starw.nii.gz: 45%|▍| 357k/788k [00:00<00:00, 1.42MB/s
sub-UCL_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/782k [00:00<?, ?B/s]
sub-UCL_acq-anat_TB1TFL.json: 0%| | 0.00/2.68k [00:00<?, ?B/s]
sub-UCL_rec-uncombined8_T2starw.nii.gz: 9%| | 66.6k/782k [00:00<00:01, 612kB/s
sub-UCL_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/847k [00:00<?, ?B/s]
sub-UCL_rec-uncombined8_T2starw.nii.gz: 30%|▎| 238k/782k [00:00<00:00, 1.25MB/s
sub-UCL_acq-anat_TB1TFL.nii.gz: 10%|▊ | 83.6k/847k [00:00<00:01, 603kB/s]
sub-UCL_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-UCL_rec-uncombined8_T2starw.nii.gz: 93%|▉| 730k/782k [00:00<00:00, 2.52MB/s
sub-UCL_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-UCL_acq-anat_TB1TFL.nii.gz: 42%|███▎ | 356k/847k [00:00<00:00, 1.40MB/s]
sub-UCL_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.76M [00:00<?, ?B/s]
sub-UCL_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.5k/3.76M [00:00<00:06, 605kB/s
sub-UCL_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-UCL_acq-coilQaSagLarge_SNR.nii.gz: 5%| | 186k/3.76M [00:00<00:04, 821kB/s]
sub-UCL_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.64M [00:00<?, ?B/s]
sub-UCL_acq-coilQaSagLarge_SNR.nii.gz: 18%|▏| 713k/3.76M [00:00<00:01, 2.61MB/s
sub-UCL_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.64M [00:00<00:04, 612
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-UCL_acq-coilQaSagLarge_SNR.nii.gz: 75%|▊| 2.83M/3.76M [00:00<00:00, 9.78MB/
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/11.7M [00:00<00:20, 591kB/s]
sub-UCL_acq-coilQaSagSmall_GFactor.nii.gz: 13%|▏| 356k/2.64M [00:00<00:01, 1.38
sub-UCL_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/90.9k [00:00<?, ?B/s]
sub-UCL_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.5k [00:00<?, ?B/s]
sub-UCL_acq-coilQaSagSmall_GFactor.nii.gz: 55%|▌| 1.46M/2.64M [00:00<00:00, 4.6
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 3%| | 373k/11.7M [00:00<00:07, 1.49MB/s]
sub-UCL_acq-famp-0.66_TB1DREAM.nii.gz: 92%|▉| 83.6k/90.9k [00:00<00:00, 611kB/s
sub-UCL_acq-famp-1.5_TB1DREAM.nii.gz: 91%|▉| 83.6k/91.5k [00:00<00:00, 590kB/s]
sub-UCL_acq-coilQaSagSmall_GFactor.nii.gz: 93%|▉| 2.45M/2.64M [00:00<00:00, 6.4
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 10%| | 1.13M/11.7M [00:00<00:02, 3.93MB/s
sub-UCL_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 23%|▏| 2.67M/11.7M [00:00<00:01, 8.18MB/s
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 59%|▌| 6.86M/11.7M [00:00<00:00, 20.0MB/s
sub-UCL_acq-famp_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-UCL_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/90.7k [00:00<?, ?B/s]
sub-UCL_acq-coilQaTra_GFactor.nii.gz: 88%|▉| 10.3M/11.7M [00:00<00:00, 25.0MB/s
sub-UCL_acq-famp_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-UCL_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/921k [00:00<?, ?B/s]
sub-UCL_acq-famp_TB1DREAM.nii.gz: 73%|███▋ | 66.5k/90.7k [00:00<00:00, 485kB/s]
sub-UCL_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL_acq-famp_TB1TFL.nii.gz: 9%|▋ | 83.6k/921k [00:00<00:01, 584kB/s]
sub-UCL_acq-famp_TB1TFL.nii.gz: 39%|███ | 356k/921k [00:00<00:00, 1.45MB/s]
sub-UCL_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/88.7k [00:00<?, ?B/s]
sub-UCL_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/81.8k [00:00<?, ?B/s]
sub-UCL_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL_acq-refv-1.5_TB1DREAM.nii.gz: 94%|▉| 83.6k/88.7k [00:00<00:00, 613kB/s]
sub-UCL_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/86.0k [00:00<?, ?B/s]
sub-UCL_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 81.8k/81.8k [00:00<00:00, 646kB/s
sub-UCL_acq-refv_TB1DREAM.nii.gz: 99%|████▉| 85.2k/86.0k [00:00<00:00, 625kB/s]
sub-UCL_acq-refv_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
✅ Finished downloading ds005090.
🧠 Please enjoy your brains.
# Define path (needs to be run in the root directory of the project)
path_data = os.path.join(os.getcwd(), "data-phantom/")
print(f"path_data: {path_data}")
path_labels = os.path.join(path_data, "derivatives", "labels")
path_qc = os.path.join(path_data, "qc")
subjects = [os.path.basename(subject_path) for subject_path in sorted(glob.glob(os.path.join(path_data, "sub-*")))]
print(f"subjects: {subjects}")
path_results = os.path.join(path_data, "derivatives", "results")
# Create results directory
os.makedirs(path_results, exist_ok=True)
path_data: /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/
subjects: ['sub-CRMBM', 'sub-MGH', 'sub-MNI', 'sub-MPI', 'sub-MSSM', 'sub-NTNU', 'sub-UCL']
Convert TFL and DREAM flip angle maps to B1+ in units of nT/V#
# Convert the TFL flip angle maps to B1+ efficiency maps [nT/V] (inspired by code from Kyle Gilbert)
# The approach consists in calculating the B1+ efficiency using a 1ms, pi-pulse at the acquisition voltage,
# then scale the efficiency by the ratio of the measured flip angle to the requested flip angle in the pulse sequence.
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp_TB1TFL.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
print(f"ref_voltage [V]: {ref_voltage} ({subject}_acq-famp_TB1TFL)")
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp_TB1TFL.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
print(f"flip angle [degrees]: {requested_fa} ({subject}_acq-famp_TB1TFL)")
# Open flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp_TB1TFL.nii.gz")
meas_fa = nii.get_fdata()
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
# Siemens maps are in units of flip angle * 10 (in degrees)
b1_map = ((meas_fa / 10) / requested_fa) * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
# Save B1 map in [T/V] as NIfTI file
nii_b1 = nib.Nifti1Image(b1_map, nii.affine, nii.header)
nib.save(nii_b1, f"{subject}_TFLTB1map.nii.gz")
ref_voltage [V]: 224.5 (sub-CRMBM_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-CRMBM_acq-famp_TB1TFL)
ref_voltage [V]: 419.5 (sub-MGH_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MGH_acq-famp_TB1TFL)
ref_voltage [V]: 464 (sub-MNI_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MNI_acq-famp_TB1TFL)
ref_voltage [V]: 408 (sub-MPI_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-MPI_acq-famp_TB1TFL)
ref_voltage [V]: 422 (sub-MSSM_acq-famp_TB1TFL)
flip angle [degrees]: 68 (sub-MSSM_acq-famp_TB1TFL)
ref_voltage [V]: 408 (sub-NTNU_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-NTNU_acq-famp_TB1TFL)
ref_voltage [V]: 270 (sub-UCL_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-UCL_acq-famp_TB1TFL)
# load DREAM FA maps acquired with different reference voltages
# threshold FA maps to 20deg < FA < 50deg
# combine FA maps by averaging non-zero estimates of FA in each pixel
voltages = ["1.5", "0.66"]
for subject in subjects:
b1_maps = []
os.chdir(os.path.join(path_data, subject, "fmap"))
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp_TB1DREAM.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Open refV flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp_TB1DREAM.nii.gz")
meas_fa = nii.get_fdata()
#thresholding
meas_fa[meas_fa < 200] = np.nan
meas_fa[meas_fa > 500] = np.nan
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp_TB1DREAM.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
#convert measured FA to percent of requested FA (note that measured FA map is in degrees * 10)
meas_fa = (meas_fa/10) / requested_fa
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
b1_map = meas_fa * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
b1_maps.append(b1_map)
for voltage in voltages:
#check if map exists
my_file = Path(f"{subject}_acq-famp-{voltage}_TB1DREAM.nii.gz")
if my_file.is_file():
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp-{voltage}_TB1DREAM.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Open flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp-{voltage}_TB1DREAM.nii.gz")
meas_fa = nii.get_fdata()
#thresholding
meas_fa[meas_fa < 200] = np.nan
meas_fa[meas_fa > 500] = np.nan
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp-{voltage}_TB1DREAM.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
#convert measured FA to percent of requested FA (note that measured FA map is in degrees * 10)
meas_fa = (meas_fa/10) / requested_fa
else:
meas_fa = np.full((nii.header).get_data_shape(),np.nan)
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
# Siemens maps are in units of flip angle * 10 (in degrees)
b1_map = meas_fa * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
b1_maps.append(b1_map)
# compute mean of non-zero values
avgB1=np.nanmean(b1_maps,axis=0)
# Save as NIfTI file
nii_avgB1 = nib.Nifti1Image(avgB1, nii.affine, nii.header)
nib.save(nii_avgB1, f"{subject}_DREAMTB1avgB1map.nii.gz")
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_12874/3268472193.py:89: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
# Split SNR data across multiple volumes. The first volume corresponds to the SoS SNR reconstruction, which is
# what we will use for SNR comparison.
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
!sct_image -i {subject}_acq-coilQaSagLarge_SNR.nii.gz -split t -o {subject}_acq-coilQaSagLarge_SNR.nii.gz
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-CRMBM_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MGH_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MGH_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MNI_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MNI_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MPI_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MPI_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MSSM_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-NTNU_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-UCL_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-UCL_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
Generate approximate mask of spinal cord#
The purpose of this section is to generate an approximate mask of the spinal cord that will be used to quantify B1+ values inside the phantom. To do this, we identified a representative human subject, overlaid it on the anthropomorphic phantom, created a few pointwise labels along the spinal cord, interpolated those points using bsplines, and then created a cylindrical mask of 10mm diameter along the generated centerline.
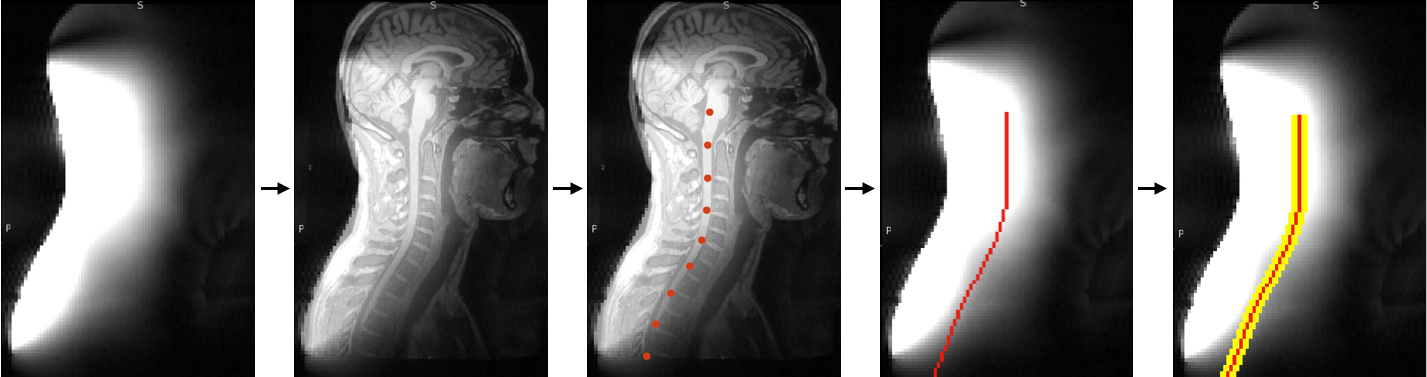
os.chdir(os.path.join(path_data, "sub-CRMBM", "fmap"))
# Create labels on TFL B1+ map along an approximate spinal cord
! sct_label_utils -i "sub-CRMBM_acq-anat_TB1TFL.nii.gz" -create 49,108,23,1:49,99,23,1:49,88,23,1:50,76,23,1:55,63,23,1:62,51,23,1:68,35,23,1:72,26,23,1 -o "label_cord.nii.gz"
# Create spinal cord centerline
! sct_get_centerline -i "label_cord.nii.gz" -method fitseg -o "centerline_spinalcord.nii.gz"
# Create mask of 10mm diameter around centerline
! sct_create_mask -i "sub-CRMBM_acq-anat_TB1TFL.nii.gz" -p centerline,"centerline_spinalcord.nii.gz" -size 10mm -f cylinder -o "sub-CRMBM_TFLTB1map_mask.nii.gz"
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM_acq-anat_TB1TFL.nii.gz -create 49,108,23,1:49,99,23,1:49,88,23,1:50,76,23,1:55,63,23,1:62,51,23,1:68,35,23,1:72,26,23,1 -o label_cord.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_get_centerline -i label_cord.nii.gz -method fitseg -o centerline_spinalcord.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_create_mask -i sub-CRMBM_acq-anat_TB1TFL.nii.gz -p centerline,centerline_spinalcord.nii.gz -size 10mm -f cylinder -o sub-CRMBM_TFLTB1map_mask.nii.gz
--
OK: centerline_spinalcord.nii.gz
Creating temporary folder (/tmp/sct_2025-03-20_02-59-15_create-mask_cv2ltr_h)
Orientation:
AIR
Dimensions:
(56, 88, 144, 1, 2.467333, 2.361111, 2.361111, 1)
Create mask...
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'uint8', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-15_create-mask_cv2ltr_h
Co-register fmap data#
The purpose of this section if to co-register phantom data across sites, to produce figures showing consistent placement of the phantom across sites, and to be able to use a single mask of the approximate spinal cord for quantitative analysis.
The co-registration uses programatically-created labels that correspond to the posterior tip of the phantom, at the inflexion point:
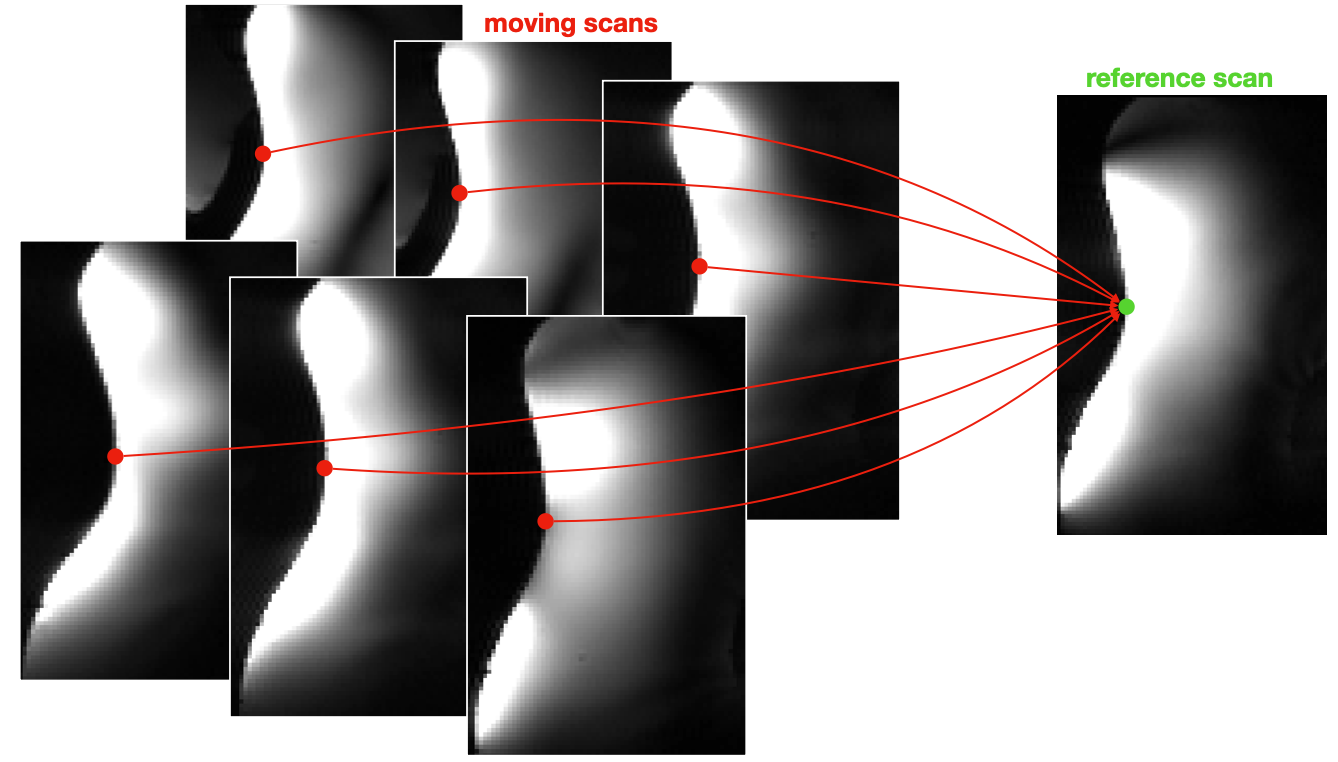
# Create dictionary of labels across sites
labels_phantom_TFL = {
'sub-CRMBM': '68,88,23',
'sub-MGH': '61,98,26',
'sub-MNI': '69,87,26',
'sub-MPI': '67,95,27',
'sub-MSSM': '73,65,28',
'sub-NTNU': '64,96,29',
'sub-UCL': '68,97,28',
}
# Define reference subject and suffix
subject_ref = 'sub-CRMBM'
suffix_ref = 'TFLTB1map'
suffixes = ["TFLTB1map", "DREAMTB1avgB1map", "acq-coilQaSagLarge_SNR_T0000"]
# Create labels for all suffixes, for all subjects
for subject in subjects:
print(f"👉 CREATE LABELS FOR: {subject}")
os.chdir(os.path.join(path_data, subject, "fmap"))
!sct_label_utils -i {subject}_{suffix_ref}.nii.gz -create {labels_phantom_TFL[subject]},1 -o {subject}_{suffix_ref}_label.nii.gz
# Register label from TFLTB1map to other metric using the qform matrix (ie: no optimization)
for suffix in [other_suffixes for other_suffixes in suffixes if other_suffixes != suffix_ref]:
!sct_register_multimodal -i {subject}_{suffix_ref}_label.nii.gz -d {subject}_{suffix}.nii.gz -identity 1
!sct_apply_transfo -i {subject}_{suffix_ref}_label.nii.gz -d {subject}_{suffix}.nii.gz -w warp_{subject}_{suffix_ref}_label2{subject}_{suffix}.nii.gz -x label -o {subject}_{suffix}_label.nii.gz
# Register other scans to the reference scan
for subject in subjects:
print(f"👉 REGISTER: {subject}")
for suffix in suffixes:
os.chdir(os.path.join(path_data, subject, "fmap"))
# Perform registration
!sct_register_multimodal -i {subject}_{suffix}.nii.gz -ilabel {subject}_{suffix}_label.nii.gz -d {path_data}{subject_ref}/fmap/{subject_ref}_{suffix}.nii.gz -dlabel {path_data}{subject_ref}/fmap/{subject_ref}_{suffix}_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
# Register TFL B1+ spinal cord mask to DREAM and SNR reference scan
os.chdir(os.path.join(path_data, subject_ref, "fmap"))
for suffix in [other_suffixes for other_suffixes in suffixes if other_suffixes != suffix_ref]:
!sct_register_multimodal -i {subject_ref}_{suffix_ref}_mask.nii.gz -d {subject_ref}_{suffix}.nii.gz -identity 1 -o {subject_ref}_{suffix}_mask.nii.gz
👉 CREATE LABELS FOR: sub-CRMBM
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM_TFLTB1map.nii.gz -create 68,88,23,1 -o sub-CRMBM_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_TFLTB1map_label.nii.gz -d sub-CRMBM_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-CRMBM_TFLTB1map_label.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_/warp_src2dest.nii.gz warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_TFLTB1map_label.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-17_register-wrapper_qrkk9ag_
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-CRMBM_TFLTB1map_label.nii.gz -d sub-CRMBM_DREAMTB1avgB1map.nii.gz -w warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_DREAMTB1avgB1map.nii.gz -x label -o sub-CRMBM_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-19_apply-transfo-3d-label_akqg98ai)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-19_apply-transfo-3d-label_akqg98ai/dilated_data.nii -o sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -t warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_DREAMTB1avgB1map.nii.gz -r sub-CRMBM_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-CRMBM_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-19_apply-transfo-3d-label_akqg98ai
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_TFLTB1map_label.nii.gz -d sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-CRMBM_TFLTB1map_label.nii.gz (88, 144, 56)
Destination ......... sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-CRMBM_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-CRMBM_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y/warp_src2dest.nii.gz warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_TFLTB1map_label.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-20_register-wrapper_4ohl842y
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-CRMBM_TFLTB1map_label.nii.gz -d sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-25_apply-transfo-3d-label_wqi_2_vu)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-25_apply-transfo-3d-label_wqi_2_vu/dilated_data.nii -o sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-CRMBM_TFLTB1map_label2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-25_apply-transfo-3d-label_wqi_2_vu
👉 CREATE LABELS FOR: sub-MGH
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH_TFLTB1map.nii.gz -create 61,98,26,1 -o sub-MGH_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH_TFLTB1map_label.nii.gz -d sub-MGH_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MGH_TFLTB1map_label.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-MGH_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad/warp_src2dest.nii.gz warp_sub-MGH_TFLTB1map_label2sub-MGH_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MGH_TFLTB1map_label2sub-MGH_DREAMTB1avgB1map.nii.gz
File created: sub-MGH_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad/warp_dest2src.nii.gz warp_sub-MGH_DREAMTB1avgB1map2sub-MGH_TFLTB1map_label.nii.gz
File created: warp_sub-MGH_DREAMTB1avgB1map2sub-MGH_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-28_register-wrapper_8s_cgpad
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MGH_TFLTB1map_label.nii.gz -d sub-MGH_DREAMTB1avgB1map.nii.gz -w warp_sub-MGH_TFLTB1map_label2sub-MGH_DREAMTB1avgB1map.nii.gz -x label -o sub-MGH_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-30_apply-transfo-3d-label_rra28q61)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-30_apply-transfo-3d-label_rra28q61/dilated_data.nii -o sub-MGH_DREAMTB1avgB1map_label.nii.gz -t warp_sub-MGH_TFLTB1map_label2sub-MGH_DREAMTB1avgB1map.nii.gz -r sub-MGH_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MGH/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-MGH_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-30_apply-transfo-3d-label_rra28q61
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH_TFLTB1map_label.nii.gz -d sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MGH_TFLTB1map_label.nii.gz (88, 144, 56)
Destination ......... sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MGH_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-MGH_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo/warp_src2dest.nii.gz warp_sub-MGH_TFLTB1map_label2sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH_TFLTB1map_label2sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-MGH_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo/warp_dest2src.nii.gz warp_sub-MGH_acq-coilQaSagLarge_SNR_T00002sub-MGH_TFLTB1map_label.nii.gz
File created: warp_sub-MGH_acq-coilQaSagLarge_SNR_T00002sub-MGH_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-31_register-wrapper_cv9hfmwo
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MGH_TFLTB1map_label.nii.gz -d sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MGH_TFLTB1map_label2sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-MGH_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-37_apply-transfo-3d-label_rkffegx0)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-37_apply-transfo-3d-label_rkffegx0/dilated_data.nii -o sub-MGH_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-MGH_TFLTB1map_label2sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MGH/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-MGH_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-37_apply-transfo-3d-label_rkffegx0
👉 CREATE LABELS FOR: sub-MNI
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI_TFLTB1map.nii.gz -create 69,87,26,1 -o sub-MNI_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI_TFLTB1map_label.nii.gz -d sub-MNI_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MNI_TFLTB1map_label.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-MNI_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6/warp_src2dest.nii.gz warp_sub-MNI_TFLTB1map_label2sub-MNI_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MNI_TFLTB1map_label2sub-MNI_DREAMTB1avgB1map.nii.gz
File created: sub-MNI_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6/warp_dest2src.nii.gz warp_sub-MNI_DREAMTB1avgB1map2sub-MNI_TFLTB1map_label.nii.gz
File created: warp_sub-MNI_DREAMTB1avgB1map2sub-MNI_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-40_register-wrapper_sbn3htq6
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MNI_TFLTB1map_label.nii.gz -d sub-MNI_DREAMTB1avgB1map.nii.gz -w warp_sub-MNI_TFLTB1map_label2sub-MNI_DREAMTB1avgB1map.nii.gz -x label -o sub-MNI_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-42_apply-transfo-3d-label_33xceiom)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-42_apply-transfo-3d-label_33xceiom/dilated_data.nii -o sub-MNI_DREAMTB1avgB1map_label.nii.gz -t warp_sub-MNI_TFLTB1map_label2sub-MNI_DREAMTB1avgB1map.nii.gz -r sub-MNI_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MNI/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-MNI_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-42_apply-transfo-3d-label_33xceiom
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI_TFLTB1map_label.nii.gz -d sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MNI_TFLTB1map_label.nii.gz (88, 144, 56)
Destination ......... sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MNI_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-MNI_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr/warp_src2dest.nii.gz warp_sub-MNI_TFLTB1map_label2sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI_TFLTB1map_label2sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-MNI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr/warp_dest2src.nii.gz warp_sub-MNI_acq-coilQaSagLarge_SNR_T00002sub-MNI_TFLTB1map_label.nii.gz
File created: warp_sub-MNI_acq-coilQaSagLarge_SNR_T00002sub-MNI_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-43_register-wrapper_pwbp1wlr
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MNI_TFLTB1map_label.nii.gz -d sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MNI_TFLTB1map_label2sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-MNI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-48_apply-transfo-3d-label_ey3qsqc6)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-48_apply-transfo-3d-label_ey3qsqc6/dilated_data.nii -o sub-MNI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-MNI_TFLTB1map_label2sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MNI/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-MNI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-48_apply-transfo-3d-label_ey3qsqc6
👉 CREATE LABELS FOR: sub-MPI
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI_TFLTB1map.nii.gz -create 67,95,27,1 -o sub-MPI_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI_TFLTB1map_label.nii.gz -d sub-MPI_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MPI_TFLTB1map_label.nii.gz (90, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-MPI_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc/warp_src2dest.nii.gz warp_sub-MPI_TFLTB1map_label2sub-MPI_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MPI_TFLTB1map_label2sub-MPI_DREAMTB1avgB1map.nii.gz
File created: sub-MPI_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc/warp_dest2src.nii.gz warp_sub-MPI_DREAMTB1avgB1map2sub-MPI_TFLTB1map_label.nii.gz
File created: warp_sub-MPI_DREAMTB1avgB1map2sub-MPI_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-52_register-wrapper_c2_lnlxc
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MPI_TFLTB1map_label.nii.gz -d sub-MPI_DREAMTB1avgB1map.nii.gz -w warp_sub-MPI_TFLTB1map_label2sub-MPI_DREAMTB1avgB1map.nii.gz -x label -o sub-MPI_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
90 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-53_apply-transfo-3d-label_07lk6opt)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_02-59-53_apply-transfo-3d-label_07lk6opt/dilated_data.nii -o sub-MPI_DREAMTB1avgB1map_label.nii.gz -t warp_sub-MPI_TFLTB1map_label2sub-MPI_DREAMTB1avgB1map.nii.gz -r sub-MPI_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MPI/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-MPI_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-53_apply-transfo-3d-label_07lk6opt
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI_TFLTB1map_label.nii.gz -d sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MPI_TFLTB1map_label.nii.gz (90, 144, 56)
Destination ......... sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MPI_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-MPI_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_/warp_src2dest.nii.gz warp_sub-MPI_TFLTB1map_label2sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI_TFLTB1map_label2sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-MPI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_/warp_dest2src.nii.gz warp_sub-MPI_acq-coilQaSagLarge_SNR_T00002sub-MPI_TFLTB1map_label.nii.gz
File created: warp_sub-MPI_acq-coilQaSagLarge_SNR_T00002sub-MPI_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-59-54_register-wrapper_yncco7y_
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MPI_TFLTB1map_label.nii.gz -d sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MPI_TFLTB1map_label2sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-MPI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
90 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-00_apply-transfo-3d-label_lfk5ogtr)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-00_apply-transfo-3d-label_lfk5ogtr/dilated_data.nii -o sub-MPI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-MPI_TFLTB1map_label2sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MPI/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-MPI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-00_apply-transfo-3d-label_lfk5ogtr
👉 CREATE LABELS FOR: sub-MSSM
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM_TFLTB1map.nii.gz -create 73,65,28,1 -o sub-MSSM_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM_TFLTB1map_label.nii.gz -d sub-MSSM_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MSSM_TFLTB1map_label.nii.gz (88, 128, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-MSSM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h/warp_src2dest.nii.gz warp_sub-MSSM_TFLTB1map_label2sub-MSSM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MSSM_TFLTB1map_label2sub-MSSM_DREAMTB1avgB1map.nii.gz
File created: sub-MSSM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h/warp_dest2src.nii.gz warp_sub-MSSM_DREAMTB1avgB1map2sub-MSSM_TFLTB1map_label.nii.gz
File created: warp_sub-MSSM_DREAMTB1avgB1map2sub-MSSM_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-03_register-wrapper_wg0pe47h
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MSSM_TFLTB1map_label.nii.gz -d sub-MSSM_DREAMTB1avgB1map.nii.gz -w warp_sub-MSSM_TFLTB1map_label2sub-MSSM_DREAMTB1avgB1map.nii.gz -x label -o sub-MSSM_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 128 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-05_apply-transfo-3d-label_wpt4y3r9)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-05_apply-transfo-3d-label_wpt4y3r9/dilated_data.nii -o sub-MSSM_DREAMTB1avgB1map_label.nii.gz -t warp_sub-MSSM_TFLTB1map_label2sub-MSSM_DREAMTB1avgB1map.nii.gz -r sub-MSSM_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MSSM/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-MSSM_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-05_apply-transfo-3d-label_wpt4y3r9
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM_TFLTB1map_label.nii.gz -d sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-MSSM_TFLTB1map_label.nii.gz (88, 128, 56)
Destination ......... sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MSSM_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze/warp_src2dest.nii.gz warp_sub-MSSM_TFLTB1map_label2sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM_TFLTB1map_label2sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-MSSM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze/warp_dest2src.nii.gz warp_sub-MSSM_acq-coilQaSagLarge_SNR_T00002sub-MSSM_TFLTB1map_label.nii.gz
File created: warp_sub-MSSM_acq-coilQaSagLarge_SNR_T00002sub-MSSM_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-06_register-wrapper_gxqoo8ze
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-MSSM_TFLTB1map_label.nii.gz -d sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MSSM_TFLTB1map_label2sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-MSSM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 128 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-11_apply-transfo-3d-label_7lq0c2ee)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-11_apply-transfo-3d-label_7lq0c2ee/dilated_data.nii -o sub-MSSM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-MSSM_TFLTB1map_label2sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MSSM/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-MSSM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-11_apply-transfo-3d-label_7lq0c2ee
👉 CREATE LABELS FOR: sub-NTNU
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU_TFLTB1map.nii.gz -create 64,96,29,1 -o sub-NTNU_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU_TFLTB1map_label.nii.gz -d sub-NTNU_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-NTNU_TFLTB1map_label.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-NTNU_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5/warp_src2dest.nii.gz warp_sub-NTNU_TFLTB1map_label2sub-NTNU_DREAMTB1avgB1map.nii.gz
File created: warp_sub-NTNU_TFLTB1map_label2sub-NTNU_DREAMTB1avgB1map.nii.gz
File created: sub-NTNU_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5/warp_dest2src.nii.gz warp_sub-NTNU_DREAMTB1avgB1map2sub-NTNU_TFLTB1map_label.nii.gz
File created: warp_sub-NTNU_DREAMTB1avgB1map2sub-NTNU_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-14_register-wrapper_ems2ecf5
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-NTNU_TFLTB1map_label.nii.gz -d sub-NTNU_DREAMTB1avgB1map.nii.gz -w warp_sub-NTNU_TFLTB1map_label2sub-NTNU_DREAMTB1avgB1map.nii.gz -x label -o sub-NTNU_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-16_apply-transfo-3d-label_zcve8j_l)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-16_apply-transfo-3d-label_zcve8j_l/dilated_data.nii -o sub-NTNU_DREAMTB1avgB1map_label.nii.gz -t warp_sub-NTNU_TFLTB1map_label2sub-NTNU_DREAMTB1avgB1map.nii.gz -r sub-NTNU_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-NTNU/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-NTNU_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-16_apply-transfo-3d-label_zcve8j_l
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU_TFLTB1map_label.nii.gz -d sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-NTNU_TFLTB1map_label.nii.gz (88, 144, 56)
Destination ......... sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-NTNU_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at/warp_src2dest.nii.gz warp_sub-NTNU_TFLTB1map_label2sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU_TFLTB1map_label2sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-NTNU_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at/warp_dest2src.nii.gz warp_sub-NTNU_acq-coilQaSagLarge_SNR_T00002sub-NTNU_TFLTB1map_label.nii.gz
File created: warp_sub-NTNU_acq-coilQaSagLarge_SNR_T00002sub-NTNU_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-17_register-wrapper_gn_xt8at
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-NTNU_TFLTB1map_label.nii.gz -d sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-NTNU_TFLTB1map_label2sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-NTNU_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-23_apply-transfo-3d-label_37yd66wz)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-23_apply-transfo-3d-label_37yd66wz/dilated_data.nii -o sub-NTNU_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-NTNU_TFLTB1map_label2sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-NTNU/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-NTNU_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-23_apply-transfo-3d-label_37yd66wz
👉 CREATE LABELS FOR: sub-UCL
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL_TFLTB1map.nii.gz -create 68,97,28,1 -o sub-UCL_TFLTB1map_label.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL_TFLTB1map_label.nii.gz -d sub-UCL_DREAMTB1avgB1map.nii.gz -identity 1
--
Input parameters:
Source .............. sub-UCL_TFLTB1map_label.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-UCL_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy/warp_src2dest.nii.gz warp_sub-UCL_TFLTB1map_label2sub-UCL_DREAMTB1avgB1map.nii.gz
File created: warp_sub-UCL_TFLTB1map_label2sub-UCL_DREAMTB1avgB1map.nii.gz
File created: sub-UCL_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy/warp_dest2src.nii.gz warp_sub-UCL_DREAMTB1avgB1map2sub-UCL_TFLTB1map_label.nii.gz
File created: warp_sub-UCL_DREAMTB1avgB1map2sub-UCL_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-26_register-wrapper_it7vvmcy
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-UCL_TFLTB1map_label.nii.gz -d sub-UCL_DREAMTB1avgB1map.nii.gz -w warp_sub-UCL_TFLTB1map_label2sub-UCL_DREAMTB1avgB1map.nii.gz -x label -o sub-UCL_DREAMTB1avgB1map_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-28_apply-transfo-3d-label_zhj71wt5)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-28_apply-transfo-3d-label_zhj71wt5/dilated_data.nii -o sub-UCL_DREAMTB1avgB1map_label.nii.gz -t warp_sub-UCL_TFLTB1map_label2sub-UCL_DREAMTB1avgB1map.nii.gz -r sub-UCL_DREAMTB1avgB1map.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-UCL/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Take the center of mass of each registered dilated labels...
File sub-UCL_DREAMTB1avgB1map_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-28_apply-transfo-3d-label_zhj71wt5
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL_TFLTB1map_label.nii.gz -d sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1
--
Input parameters:
Source .............. sub-UCL_TFLTB1map_label.nii.gz (88, 144, 56)
Destination ......... sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-UCL_TFLTB1map_label_reg.nii.gz already exists. Deleting it..
File created: sub-UCL_TFLTB1map_label_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc/warp_src2dest.nii.gz warp_sub-UCL_TFLTB1map_label2sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL_TFLTB1map_label2sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-UCL_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc/warp_dest2src.nii.gz warp_sub-UCL_acq-coilQaSagLarge_SNR_T00002sub-UCL_TFLTB1map_label.nii.gz
File created: warp_sub-UCL_acq-coilQaSagLarge_SNR_T00002sub-UCL_TFLTB1map_label.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-29_register-wrapper_9w3qrvpc
Finished! Elapsed time: 4s
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i sub-UCL_TFLTB1map_label.nii.gz -d sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-UCL_TFLTB1map_label2sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -x label -o sub-UCL_acq-coilQaSagLarge_SNR_T0000_label.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
88 x 144 x 56 x 1
Apply transformation...
Dilate labels before warping...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-34_apply-transfo-3d-label_nnc1avqf)
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i /tmp/sct_2025-03-20_03-00-34_apply-transfo-3d-label_nnc1avqf/dilated_data.nii -o sub-UCL_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -t warp_sub-UCL_TFLTB1map_label2sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-UCL/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Take the center of mass of each registered dilated labels...
File sub-UCL_acq-coilQaSagLarge_SNR_T0000_label.nii.gz already exists. Will overwrite it.
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-34_apply-transfo-3d-label_nnc1avqf
👉 REGISTER: sub-CRMBM
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_TFLTB1map.nii.gz -ilabel sub-CRMBM_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-11.087461948394775, 50.14370059967041, -3.270395278930664]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 45
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[0.00000000e+00 1.66893006e-06 0.00000000e+00]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_TFLTB1map_src_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg/warp_src2dest.nii.gz warp_sub-CRMBM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_dest_reg.nii.gz
File warp_sub-CRMBM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz already exists. Deleting it..
mv /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-36_register-wrapper_sawc8ecg
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_DREAMTB1avgB1map.nii.gz -ilabel sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 1
Function evaluations: 22
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[0. 0. 0.]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_DREAMTB1avgB1map_src_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw/warp_src2dest.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_dest_reg.nii.gz
File warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz already exists. Deleting it..
mv /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-39_register-wrapper_r7oh_0rw
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-11.117252349853516, 50.47705077917857, -2.909301755652923]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 62
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[0.00000000e+00 0.00000000e+00 3.75352307e-09]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_src_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9/warp_src2dest.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_dest_reg.nii.gz
File warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Deleting it..
mv /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-41_register-wrapper_62lhvyl9
Finished! Elapsed time: 7s
👉 REGISTER: sub-MGH
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH_TFLTB1map.nii.gz -ilabel sub-MGH_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MGH_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-1.9035463333129883, 10.127162218093872, 35.712753772735596]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[ 9.18391562 -40.01653671 38.98314905]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m/warp_src2dest.nii.gz warp_sub-MGH_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-MGH_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-MGH_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-MGH_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-49_register-wrapper_iz_cwy_m
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH_DREAMTB1avgB1map.nii.gz -ilabel sub-MGH_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MGH_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-2.8459887504577637, 9.98828125, 34.462745666503906]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[ 8.27126169 -40.98875427 37.87203217]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MGH_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-MGH_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh/warp_src2dest.nii.gz warp_sub-MGH_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MGH_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-MGH_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-MGH_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-52_register-wrapper_ejlvsxhh
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-MGH_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-2.645986557006836, 10.48828125, 36.462738037109375]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[ 8.47126579 -39.98876953 39.3720398 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MGH_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MGH_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep/warp_src2dest.nii.gz warp_sub-MGH_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MGH_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-00-54_register-wrapper_9yo_ijep
Finished! Elapsed time: 7s
👉 REGISTER: sub-MNI
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI_TFLTB1map.nii.gz -ilabel sub-MNI_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MNI_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-5.0127153396606445, 12.580960512161255, 37.777780294418335]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[ 6.07474661 -37.56273842 41.04817557]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj/warp_src2dest.nii.gz warp_sub-MNI_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-MNI_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-MNI_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-MNI_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-02_register-wrapper_eon2dghj
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI_DREAMTB1avgB1map.nii.gz -ilabel sub-MNI_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MNI_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-4.612692832946777, 12.250770568847656, 38.44549560546875]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[ 6.50455761 -38.72626495 41.8547821 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MNI_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-MNI_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph/warp_src2dest.nii.gz warp_sub-MNI_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MNI_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-MNI_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-MNI_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-05_register-wrapper_08dpx_ph
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-MNI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-4.612680435180664, 12.75076293739636, 38.44549560754753]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[ 6.50457191 -37.72628784 41.35479737]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MNI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MNI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw/warp_src2dest.nii.gz warp_sub-MNI_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MNI_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-07_register-wrapper_n2da9wuw
Finished! Elapsed time: 7s
👉 REGISTER: sub-MPI
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI_TFLTB1map.nii.gz -ilabel sub-MPI_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MPI_TFLTB1map.nii.gz (90, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-8.676316738128662, 5.191625118255615, 34.30185270309448]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[ 2.41114521 -44.95207381 37.57224798]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_/warp_src2dest.nii.gz warp_sub-MPI_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-MPI_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-MPI_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-MPI_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-15_register-wrapper_mn0oki7_
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI_DREAMTB1avgB1map.nii.gz -ilabel sub-MPI_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MPI_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-9.586259841918945, 5.7471771240234375, 35.135169982910156]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[ 1.5309906 -45.2298584 38.54445648]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MPI_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-MPI_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii/warp_src2dest.nii.gz warp_sub-MPI_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MPI_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-MPI_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-MPI_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-18_register-wrapper_dkaohuii
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-MPI_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-9.486261367797852, 5.7471923828125, 34.635162353515625]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[ 1.63099098 -44.7298584 37.54446411]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MPI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MPI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r/warp_src2dest.nii.gz warp_sub-MPI_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MPI_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-20_register-wrapper_h3ddsh7r
Finished! Elapsed time: 7s
👉 REGISTER: sub-MSSM
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM_TFLTB1map.nii.gz -ilabel sub-MSSM_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MSSM_TFLTB1map.nii.gz (88, 128, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.5, 40.5, 5.0]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[13.58746195 -9.64369893 8.27039528]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4/warp_src2dest.nii.gz warp_sub-MSSM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-MSSM_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-MSSM_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-MSSM_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-28_register-wrapper_8ulcdie4
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM_DREAMTB1avgB1map.nii.gz -ilabel sub-MSSM_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MSSM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[3.5, 41.0, 5.0]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[14.61725044 -9.97703552 8.4092865 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MSSM_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf/warp_src2dest.nii.gz warp_sub-MSSM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MSSM_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-MSSM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-MSSM_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-31_register-wrapper_gi8e4shf
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-MSSM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[3.5999984741210938, 40.50000007937501, 4.499999908011118]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[14.71725082 -9.9770507 7.40930167]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MSSM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03/warp_src2dest.nii.gz warp_sub-MSSM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-MSSM_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-33_register-wrapper_t8vp1p03
Finished! Elapsed time: 7s
👉 REGISTER: sub-NTNU
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU_TFLTB1map.nii.gz -ilabel sub-NTNU_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-NTNU_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[3.7121474742889404, 5.1349334716796875, 27.99675750732422]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[ 14.79960942 -45.00876546 31.26715279]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz/warp_src2dest.nii.gz warp_sub-NTNU_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-NTNU_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-NTNU_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-NTNU_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-41_register-wrapper_k7bucnsz
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU_DREAMTB1avgB1map.nii.gz -ilabel sub-NTNU_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-NTNU_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.867208480834961, 5.4127197265625, 28.968994140625]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[ 13.98445892 -45.5643158 32.37828064]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-NTNU_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq/warp_src2dest.nii.gz warp_sub-NTNU_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-NTNU_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-NTNU_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-NTNU_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-44_register-wrapper_44bauieq
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-NTNU_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.7672119140625, 4.4127197265625, 27.468994140625]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[ 13.88446426 -46.06433105 30.3782959 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-NTNU_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98/warp_src2dest.nii.gz warp_sub-NTNU_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-NTNU_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-46_register-wrapper_yvcs1t98
Finished! Elapsed time: 7s
👉 REGISTER: sub-UCL
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL_TFLTB1map.nii.gz -ilabel sub-UCL_TFLTB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-UCL_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[1.2494821548461914, 49.14424228668213, 24.081058740615845]]
Labels dest: [[-11.087461948394775, 50.143698930740356, -3.270395278930664]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.08746195 50.14369893 -3.27039528]
Translation:
[[12.3369441 -0.99945664 27.35145402]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh/warp_src2dest.nii.gz warp_sub-UCL_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: warp_sub-UCL_TFLTB1map2sub-CRMBM_TFLTB1map.nii.gz
File created: sub-CRMBM_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh/warp_dest2src.nii.gz warp_sub-CRMBM_TFLTB1map2sub-UCL_TFLTB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map2sub-UCL_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-54_register-wrapper_q329e7hh
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL_DREAMTB1avgB1map.nii.gz -ilabel sub-UCL_DREAMTB1avgB1map_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-UCL_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[0.36720848083496094, 49.977577209472656, 25.192169189453125]]
Labels dest: [[-11.117250442504883, 50.97703552246094, -3.4092864990234375]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725044 50.97703552 -3.4092865 ]
Translation:
[[11.48445892 -0.99945831 28.60145569]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-UCL_DREAMTB1avgB1map_reg.nii.gz already exists. Deleting it..
File created: sub-UCL_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2/warp_src2dest.nii.gz warp_sub-UCL_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-UCL_DREAMTB1avgB1map2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-UCL_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-UCL_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-57_register-wrapper_ol37frp2
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz -ilabel sub-UCL_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -d /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -dlabel /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_label.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=im,algo=syn,iter=0
--
Input parameters:
Source .............. sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Destination ......... /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[0.36721134185791016, 49.477569580078125, 24.192169189453125]]
Labels dest: [[-11.117252349853516, 50.47705077917857, -2.9093017594064463]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-11.11725235 50.47705078 -2.90930176]
Translation:
[[11.48446369 -0.9994812 27.10147095]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_RPI.nii,src_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-UCL_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-UCL_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r/warp_src2dest.nii.gz warp_sub-UCL_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-UCL_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-01-59_register-wrapper__yeu2c4r
Finished! Elapsed time: 7s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_TFLTB1map_mask.nii.gz -d sub-CRMBM_DREAMTB1avgB1map.nii.gz -identity 1 -o sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz
--
Input parameters:
Source .............. sub-CRMBM_TFLTB1map_mask.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... sub-CRMBM_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name ......... sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz
mv /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44/warp_src2dest.nii.gz warp_sub-CRMBM_TFLTB1map_mask2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM_TFLTB1map_mask2sub-CRMBM_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM_DREAMTB1avgB1map_mask_inv.nii.gz
mv /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44/warp_dest2src.nii.gz warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_TFLTB1map_mask.nii.gz
File created: warp_sub-CRMBM_DREAMTB1avgB1map2sub-CRMBM_TFLTB1map_mask.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-02-07_register-wrapper_a8jhao44
Finished! Elapsed time: 1s
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM_TFLTB1map_mask.nii.gz -d sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -identity 1 -o sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz
--
Input parameters:
Source .............. sub-CRMBM_TFLTB1map_mask.nii.gz (88, 144, 56)
Destination ......... sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz (512, 512, 13)
Init transfo ........
Mask ................
Output name ......... sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz
mv /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7/warp_src2dest.nii.gz warp_sub-CRMBM_TFLTB1map_mask2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM_TFLTB1map_mask2sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask_inv.nii.gz
mv /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7/warp_dest2src.nii.gz warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_TFLTB1map_mask.nii.gz
File created: warp_sub-CRMBM_acq-coilQaSagLarge_SNR_T00002sub-CRMBM_TFLTB1map_mask.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_03-02-09_register-wrapper_s2wdg6i7
Finished! Elapsed time: 4s
# Generate syntax to open registered data in FSLeyes. This should be run from within the `data-phantom/` folder.
for suffix in suffixes:
print(f"\n👉 CHECKING REGISTRATION FOR: {suffix}\n")
cmd = f"fsleyes sub-CRMBM/fmap/sub-CRMBM_{suffix}.nii.gz"
for subject in [other_subjects for other_subjects in subjects if other_subjects != subject_ref]:
file_reg = os.path.join(path_data, subject, "fmap", f"{subject}_{suffix}_reg.nii.gz")
cmd = f"{cmd} {file_reg}"
# Add mask with color and opacity
cmd = f"{cmd} sub-CRMBM/fmap/sub-CRMBM_{suffix}_mask.nii.gz --cmap yellow -a 30"
print(cmd+" &")
# TODO: undisplay all scans but the first
👉 CHECKING REGISTRATION FOR: TFLTB1map
fsleyes sub-CRMBM/fmap/sub-CRMBM_TFLTB1map.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MGH/fmap/sub-MGH_TFLTB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MNI/fmap/sub-MNI_TFLTB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MPI/fmap/sub-MPI_TFLTB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MSSM/fmap/sub-MSSM_TFLTB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-NTNU/fmap/sub-NTNU_TFLTB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-UCL/fmap/sub-UCL_TFLTB1map_reg.nii.gz sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz --cmap yellow -a 30 &
👉 CHECKING REGISTRATION FOR: DREAMTB1avgB1map
fsleyes sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MGH/fmap/sub-MGH_DREAMTB1avgB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MNI/fmap/sub-MNI_DREAMTB1avgB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MPI/fmap/sub-MPI_DREAMTB1avgB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MSSM/fmap/sub-MSSM_DREAMTB1avgB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-NTNU/fmap/sub-NTNU_DREAMTB1avgB1map_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-UCL/fmap/sub-UCL_DREAMTB1avgB1map_reg.nii.gz sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz --cmap yellow -a 30 &
👉 CHECKING REGISTRATION FOR: acq-coilQaSagLarge_SNR_T0000
fsleyes sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MGH/fmap/sub-MGH_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MNI/fmap/sub-MNI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MPI/fmap/sub-MPI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-MSSM/fmap/sub-MSSM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-NTNU/fmap/sub-NTNU_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/sub-UCL/fmap/sub-UCL_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz --cmap yellow -a 30 &
Extract values in mask and generate figures#
# Extract B1+ and SNR along the spinal cord mask and save data to CSV files
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
fname_result_TFLb1plus = os.path.join(path_results, f"{subject}_TFLTB1map.csv")
fname_result_DREAMb1plus = os.path.join(path_results, f"{subject}_DREAMTB1avgB1map.csv")
fname_result_SNR = os.path.join(path_results, f"{subject}_acq-coilQaSagLarge_SNR_T0000.csv")
# For sites other than sub-CRMBM, use the registered qMRI map for metrics extraction
if subject=='sub-CRMBM':
file_suffix = ''
else:
file_suffix = '_reg'
# Extract metrics within mask defined in the sub-CRMBM space
!sct_extract_metric -i {subject}_TFLTB1map{file_suffix}.nii.gz -f ../../{subject_ref}/fmap/{subject_ref}_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o "{fname_result_TFLb1plus}"
!sct_extract_metric -i {subject}_DREAMTB1avgB1map{file_suffix}.nii.gz -f ../../{subject_ref}/fmap/{subject_ref}_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o "{fname_result_DREAMb1plus}"
!sct_extract_metric -i {subject}_acq-coilQaSagLarge_SNR_T0000{file_suffix}.nii.gz -f ../../{subject_ref}/fmap/{subject_ref}_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o "{fname_result_SNR}"
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM_TFLTB1map.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM_DREAMTB1avgB1map.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MGH_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MNI_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MPI_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-MSSM_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-NTNU_acq-coilQaSagLarge_SNR_T0000.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL_TFLTB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz -method wa -z 19:127 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_TFLTB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_TFLTB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL_DREAMTB1avgB1map_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz -method wa -z 14:62 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_DREAMTB1avgB1map.csv
--
Load metric image...
Estimation for label: sub-CRMBM_DREAMTB1avgB1map_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -f ../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz -method wa -z 151:368 -perslice 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_acq-coilQaSagLarge_SNR_T0000.csv
--
Load metric image...
Estimation for label: sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-phantom/derivatives/results/sub-UCL_acq-coilQaSagLarge_SNR_T0000.csv
Correct SNR using B1+ values#
SNR depends on the effective flip angle at each voxel, which is influenced by the B1+ efficiency and the transmit homogeneity. Therefore, if we want to report SNR values independently of the actual flip angle played out, we need to correct the SNR maps. To do so, we divide the reconstructed SNR maps by \(\sin(\text{FA}_{\text{gre, meas}})\), where \(\text{FA}_{\text{gre, meas}}\) is the actual flip angle map for the GRE SNR scan. By normalizing, we extrapolate to the SNR value that we would achieve with \( \text{FA} = 90^\circ \):
\(\text{FA}_{\text{gre, meas}}\) is computed from the TFL B1+ scan as follows:
The ratio \(\frac{\text{FA}_{\text{TFL, meas}}}{\text{FA}_{\text{TFL, requested}}}\) is obtained from the TFL B1+ efficiency values, which were previously extracted slicewise and saved as a CSV file.
Before doing this correction, given that the B1+ and SNR maps don’t have the same spatial resolution, we need to interpolate the B1+ array to match its length with that of the SNR array.
Reference: https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.27695
for subject in subjects:
fname_result_TFLb1plus = os.path.join(path_results, f"{subject}_TFLTB1map.csv")
fname_result_SNR = os.path.join(path_results, f"{subject}_acq-coilQaSagLarge_SNR_T0000.csv")
fname_result_SNR_corrected = os.path.join(path_results, f"{subject}_acq-coilQaSagLarge_SNR_T0000_corrected.csv")
# INTERPOLATE B1+ MAP TO MATCH THE LENGTH OF THE SNR DATA
# =======================================================
# Read B1 values
df_b1 = pd.read_csv(fname_result_TFLb1plus)
b1_map = df_b1["WA()"].values
# Read SNR values
df_snr = pd.read_csv(fname_result_SNR)
snr = df_snr["WA()"].values
# Create interpolation function based on the length of snr
x = np.linspace(0, len(b1_map) - 1, len(b1_map)) # x values for original data points
f = interp1d(x, b1_map, kind='linear')
# Generate new x values to match the length of the `snr` array
x_new = np.linspace(0, len(b1_map) - 1, len(snr))
# Interpolate to get the new y values with the length of `snr`
b1_map_interp = f(x_new)
# CORRECT SNR VALUES FOR B1+ INHOMOGENEITY
# ========================================
# Fetch the reference voltage for the TFL scan
# TODO: create a function for this
with open(os.path.join(path_data, subject, "fmap", f"{subject}_acq-famp_TB1TFL.json"), "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Fetch the flip angle for the TFL scan
with open(os.path.join(path_data, subject, "fmap", f"{subject}_acq-famp_TB1TFL.json"), "r") as f:
metadata = json.load(f)
requested_fa_tfl = metadata.get("FlipAngle", "N/A")
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# measured-to-requested flip angle using B1 values (in nT/V)
meas_to_requested_fa = 10 * b1_map_interp * GAMMA * 1e-3 * voltage_at_socket / (np.pi * 1e9)
# Fetch the requested flip angle for the SNR GRE scan
with open(os.path.join(path_data, subject, "fmap", f"{subject}_acq-coilQaSagLarge_SNR.json"), "r") as f:
metadata = json.load(f)
requested_fa_gre = metadata.get("FlipAngle", "N/A")
# Correct the SNR values for B1+ inhomogeneity
snr_corrected = snr / np.sin(np.deg2rad(requested_fa_gre * meas_to_requested_fa))
# Save corrected SNR values to CSV
df_snr_corrected = pd.DataFrame(snr_corrected)
# Insert 'WA()' as the first value in the DataFrame
df_snr_corrected.loc[-1] = ['WA()'] + [None] * (df_snr_corrected.shape[1] - 1) # Prepend 'WA()' to the first column
df_snr_corrected.index = df_snr_corrected.index + 1 # Shift index
df_snr_corrected = df_snr_corrected.sort_index() # Sort index to move the new row to the top
# Save corrected SNR values to CSV
df_snr_corrected.to_csv(fname_result_SNR_corrected, index=False, header=False)
Generate figures#
g-factor maps#
# Compute the average and maximum 1/g factor in a central ROI
# Display 1/g factor maps and T2w (coil-combined) maps for each site (MSSM excluded, see: https://github.com/spinal-cord-7t/coil-qc-code/issues/61)
sites = ["CRMBM", "MNI", "MPI", "UCL", "MGH", "NTNU"] # MSSM excluded, see: https://github.com/spinal-cord-7t/coil-qc-code/issues/61
site_colors = ['cornflowerblue', 'firebrick', 'limegreen', 'royalblue', 'darkred', 'green']
# map types
map_types = ["acq-coilQaSagSmall_GFactor", "acq-coilQaTra_GFactor", "T2starw"]
# legend types
legend_types = ["[1/g]", "[1/g]", "[a.u.]"]
mean_gfac = {}
for map_type, legend_type in zip(map_types,legend_types):
# Create a figure with multiple subplots
fig, axes = plt.subplots(2, 3, figsize=(10, 8))
font_size = 12
axes=axes.flatten()
for i,site in enumerate(sites):
# T2starw
if map_type=="T2starw":
os.chdir(os.path.join(path_data, f"sub-{site}", "anat"))
map=nib.load(f"sub-{site}_{map_type}.nii.gz")
data=map.get_fdata()[:,:,round(map.get_fdata().shape[2]/2)]
# g-factor maps
else:
os.chdir(os.path.join(path_data, f"sub-{site}", "fmap"))
map=nib.load(f"sub-{site}_{map_type}.nii.gz")
if map_type=="acq-coilQaSagSmall_GFactor":
data=(map.get_fdata()[64:191,64:191,round(map.get_fdata().shape[2]/2),5])/1000
gfac_data=(map.get_fdata()[round(map.get_fdata().shape[0]/2)-10:round(map.get_fdata().shape[0]/2)+10,round(map.get_fdata().shape[1]/2)-10:round(map.get_fdata().shape[1]/2)+10,round(map.get_fdata().shape[2]/2),5])
else:
data=(map.get_fdata()[192:574,192:574,round(map.get_fdata().shape[2]/2),0])/1000
gfac_data=(map.get_fdata()[round(map.get_fdata().shape[0]/2)-30:round(map.get_fdata().shape[0]/2)+30,round(map.get_fdata().shape[1]/2)-30:round(map.get_fdata().shape[1]/2)+30,round(map.get_fdata().shape[2]/2),5])
mean_gfac[site]=np.nanmean(gfac_data)/1000
# Plot
splot=axes[i]
dynmin = 0
if map_type=="T2starw":
dynmax = 2100
axes[-1].axis('off')
else:
dynmax = 1
axes[-1].axis('off')
splot.text(0, 3, r'mean 1/g = '+str(round(mean_gfac[site],3)), size=14)
if map_type=="acq-coilQaSagSmall_GFactor":
x = [data.shape[0]/2-10, data.shape[1]/2-10]
y = [data.shape[0]/2-10, data.shape[1]/2+10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2-10, data.shape[1]/2+10]
y = [data.shape[0]/2+10, data.shape[1]/2+10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+10, data.shape[1]/2+10]
y = [data.shape[0]/2+10, data.shape[1]/2-10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+10, data.shape[1]/2-10]
y = [data.shape[0]/2-10, data.shape[1]/2-10]
splot.plot(x, y, color="black", linewidth=2)
else:
x = [data.shape[0]/2-30, data.shape[1]/2-30]
y = [data.shape[0]/2-30, data.shape[1]/2+30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2-30, data.shape[1]/2+30]
y = [data.shape[0]/2+30, data.shape[1]/2+30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+30, data.shape[1]/2+30]
y = [data.shape[0]/2+30, data.shape[1]/2-30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+30, data.shape[1]/2-30]
y = [data.shape[0]/2-30, data.shape[1]/2-30]
splot.plot(x, y, color="black", linewidth=2)
im = splot.imshow((data.T), cmap='viridis', origin='lower',vmin=dynmin,vmax=dynmax)
splot.set_title(site, size=font_size, color=site_colors[i])
splot.axis('off')
plt.tight_layout()
# Colorbar
# Assume that the colorbar should start at the bottom of the lower row of subplots and
# extend to the top of the upper row of subplots
cbar_bottom = 0.2 # This might need adjustment
cbar_height = 0.6 # This represents the total height of both rows of subplots
cbar_dist = 1.01
cbar_ax = fig.add_axes([cbar_dist, cbar_bottom, 0.03, cbar_height])
cbar = plt.colorbar(im, cax=cbar_ax)
cbar_ax.set_title(legend_type, size=12)
plt.show()
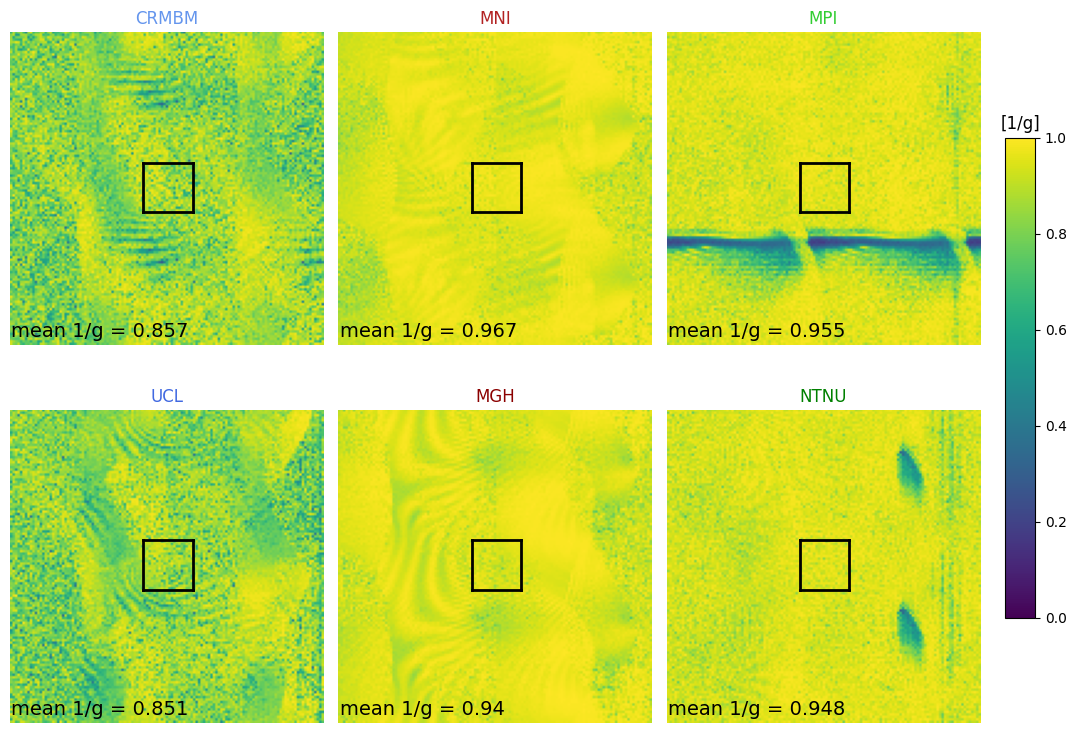
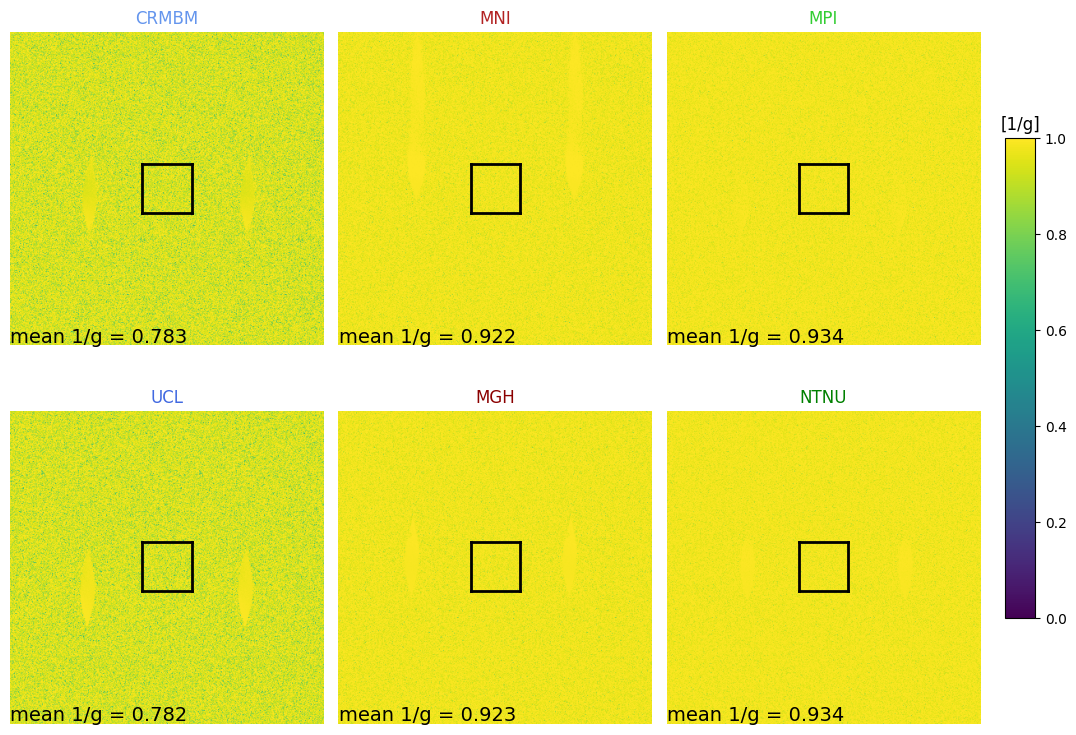
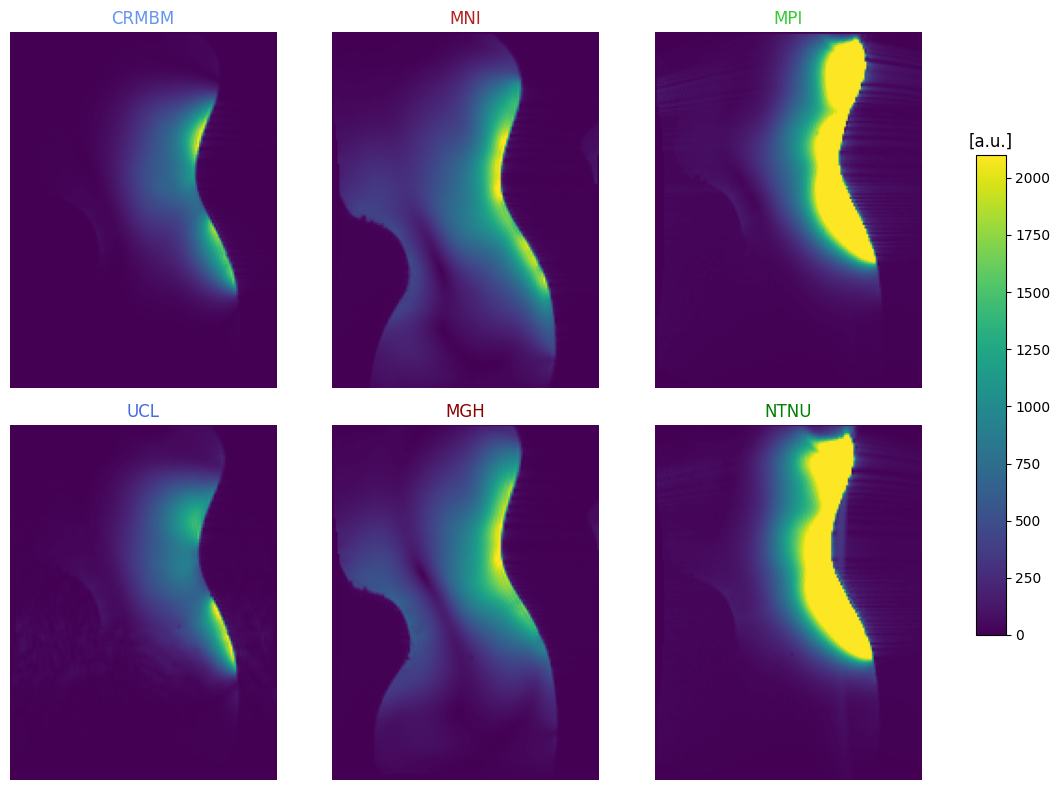
B1+ and SNR maps#
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
colors = ['cornflowerblue', 'royalblue', 'firebrick', 'darkred', 'limegreen', 'green', 'mediumpurple']
# map types
map_types = ["TFLTB1map", "DREAMTB1avgB1map", "acq-coilQaSagLarge_SNR_T0000"]
# Define the dictionary with ranges for each map type
# NIfTI voxel location: (xmin, xmax, ymin, ymax, zdata, zmask)
range_dict = {
"TFLTB1map": (19, 83, 21, 122, 23, 23),
"DREAMTB1avgB1map": (18, 79, 14, 62, 5, 5),
"acq-coilQaSagLarge_SNR_T0000": (217, 317, 176, 335, 6, 6),
}
# legend types
legend_types = ["B1+ [nT/V]", "B1+ [nT/V]", "SNR [a.u.]"]
for map_type, legend_type in zip(map_types,legend_types):
# Create a figure with multiple subplots
fig, axes = plt.subplots(2, 4, figsize=(10, 8))
font_size = 12
axes=axes.flatten()
for i,site in enumerate(sites):
# Load data
os.chdir(os.path.join(path_data, f"sub-{site}", "fmap"))
if site=='CRMBM':
map=nib.load(f"sub-{site}_{map_type}.nii.gz")
if map_type=="TFLTB1map":
mask=nib.load(f"sub-CRMBM_TFLTB1map_mask.nii.gz")
elif map_type=="DREAMTB1avgB1map":
mask=nib.load(f"sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz")
else:
mask=nib.load(f"sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz")
else:
map=nib.load(f"sub-{site}_{map_type}_reg.nii.gz")
if map_type=="TFLTB1map":
mask=nib.load(f"../../sub-CRMBM/fmap/sub-CRMBM_TFLTB1map_mask.nii.gz")
elif map_type=="DREAMTB1avgB1map":
mask=nib.load(f"../../sub-CRMBM/fmap/sub-CRMBM_DREAMTB1avgB1map_mask.nii.gz")
else:
mask=nib.load(f"../../sub-CRMBM/fmap/sub-CRMBM_acq-coilQaSagLarge_SNR_T0000_mask.nii.gz")
# Crop data for display
data = map.get_fdata()[
range_dict[map_type][0]:range_dict[map_type][1],
range_dict[map_type][2]:range_dict[map_type][3],
range_dict[map_type][4]]
mask_data = mask.get_fdata()[
range_dict[map_type][0]:range_dict[map_type][1],
range_dict[map_type][2]:range_dict[map_type][3],
range_dict[map_type][5]]
# Binarize mask for display purpose
mask_data = (mask_data > 0.5).astype(int)
# Plot
dynmin = 0
if map_type=="acq-coilQaSagLarge_SNR_T0000":
dynmax = 310
else:
dynmax = 75
splot=axes[i]
im1 = splot.imshow((data.T), cmap='viridis', origin='lower',vmin=dynmin,vmax=dynmax)
# im2 = splot.imshow((mask_data.T), 'gray', origin='lower', alpha=0.2)
splot.contour(mask_data.T, levels=[0.5], colors='white', linewidths=0.5)
splot.set_title(site, size=font_size,c=colors[i])
splot.axis('off')
# Display mean signal within mask for each site in an additional sub-plot
for site,color in zip(sites,colors):
# Read CSV file
if map_type == "acq-coilQaSagLarge_SNR_T0000":
file_csv = os.path.join(path_results, f"sub-{site}_{map_type}_corrected.csv")
else:
file_csv = os.path.join(path_results, f"sub-{site}_{map_type}.csv")
df = pd.read_csv(file_csv)
data = df['WA()'].dropna()
# Smooth data
data = uniform_filter1d(data, 10, mode='nearest')
# Display plot
axes[-1].plot(data, label=site, c=color)
axes[-1].legend(fontsize=10, loc='center left', bbox_to_anchor=(1.2, 0.5))
axes[-1].yaxis.set_label_position("right")
axes[-1].yaxis.tick_right()
axes[-1].set_xticks([]) # Remove x-axis ticks
axes[-1].set_xlabel("← superior inferior →", fontsize=12, loc='center')
# axes[-1].set_yticks([]) # Remove y-axis ticks
axes[-1].invert_xaxis() # Reverse the x-axis
axes[-1].yaxis.grid(True, color='grey', linestyle='-', linewidth=0.5)
# Reduce the space between the rows of subplots
fig.subplots_adjust(hspace=-0.2) # Adjust this value as needed to reduce spacing
# Adjust figure subplots
plt.tight_layout()
box = axes[-1].get_position()
new_height = box.height * 0.76 # Reduce the height of the second subplot by 50%
axes[-1].set_position([box.x0, box.y0 + 0.068, box.width, new_height])
# Colorbar
cbar_bottom = 0.545 # This might need adjustment
cbar_height = 0.37 # This represents the total height of both rows of subplots
if (map_type == "TFLTB1map" or map_type == "DREAMTB1avgB1map" or map_type == "acq-coilQaSagLarge_SNR_T0000"):
cbar_dist = -0.1
else:
cbar_dist = 1.01
cbar_ax = fig.add_axes([0.9, cbar_bottom, 0.02, cbar_height])
cbar = plt.colorbar(im1, cax=cbar_ax)
cbar_ax.set_title(legend_type, size=10)
plt.show()
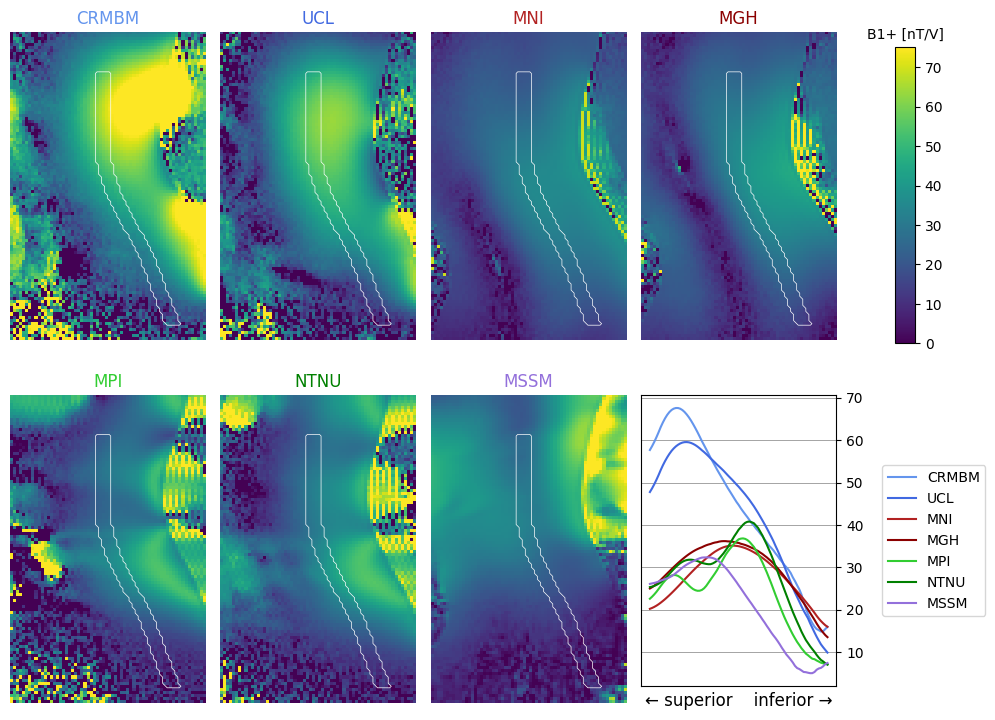
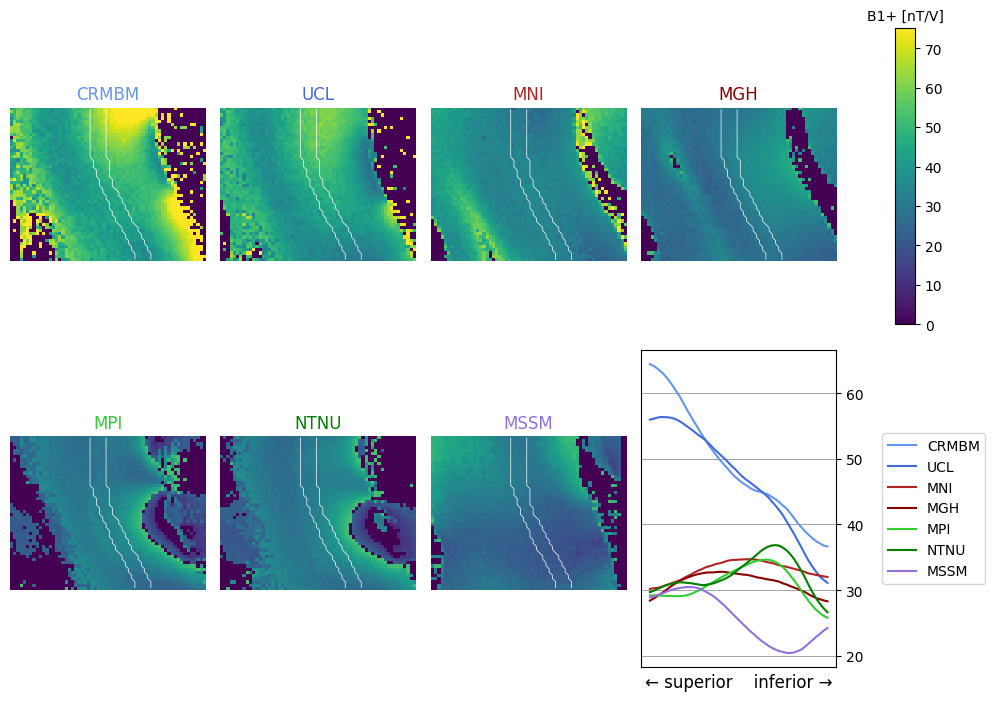
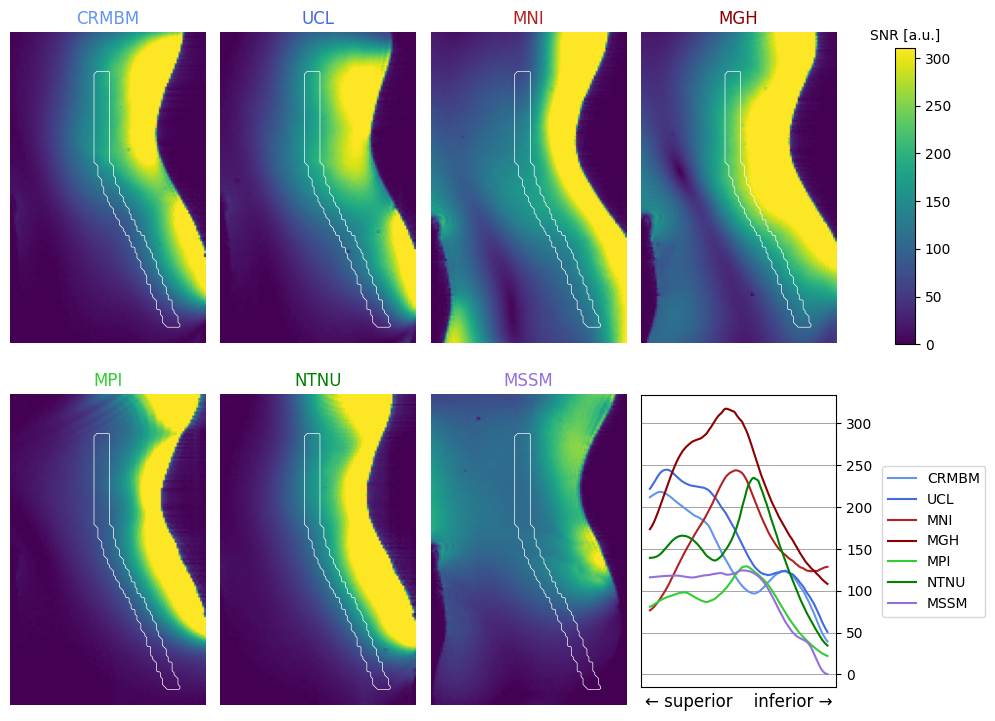
Individual coil channels#
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
for i,site in enumerate(sites):
gre_files=sorted(glob.glob(os.path.join(path_data, f"sub-{site}", "anat", '*uncombined*.nii.gz')))
#Tiled figure in a five-row layout
rows=int(np.ceil(len(gre_files)/4))
cols=int(np.ceil(len(gre_files)/rows))
fig=plt.figure(figsize=(15, 20))
ax = fig.subplots(rows,cols,squeeze=True)
for row in range(rows):
for col in range(cols):
i = row*cols+col
if i < len(gre_files):
#read in files
data_to_plot=(nib.load(gre_files[i])).get_fdata() #load in nifti object, get only image data
data_to_plot=np.rot90(data_to_plot[:,:,int(np.floor(data_to_plot.shape[2]/2))]) #central slice
ax[row,col].imshow(data_to_plot,cmap=plt.cm.gray,clim=[0, 300])
ax[row,col].text(0.5, 0.05, 'Rx channel : ' + str(i+1),horizontalalignment='center', transform=ax[row,col].transAxes,color='white',fontsize=17)
ax[row,col].axis('off')
plt.axis('off')
plt.subplots_adjust(hspace=0,wspace=0)
fig.suptitle(site, fontsize=20, y=0.9)
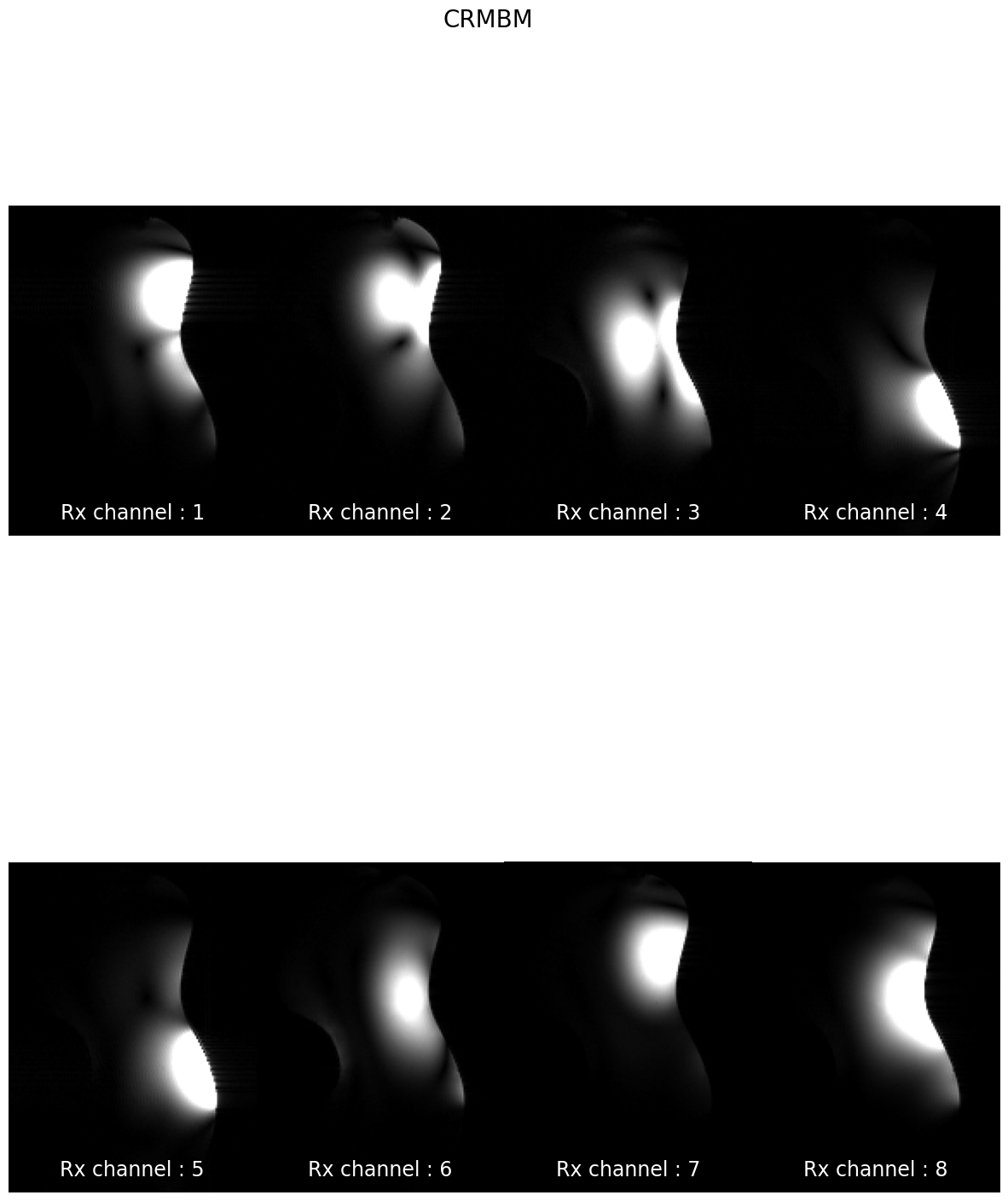
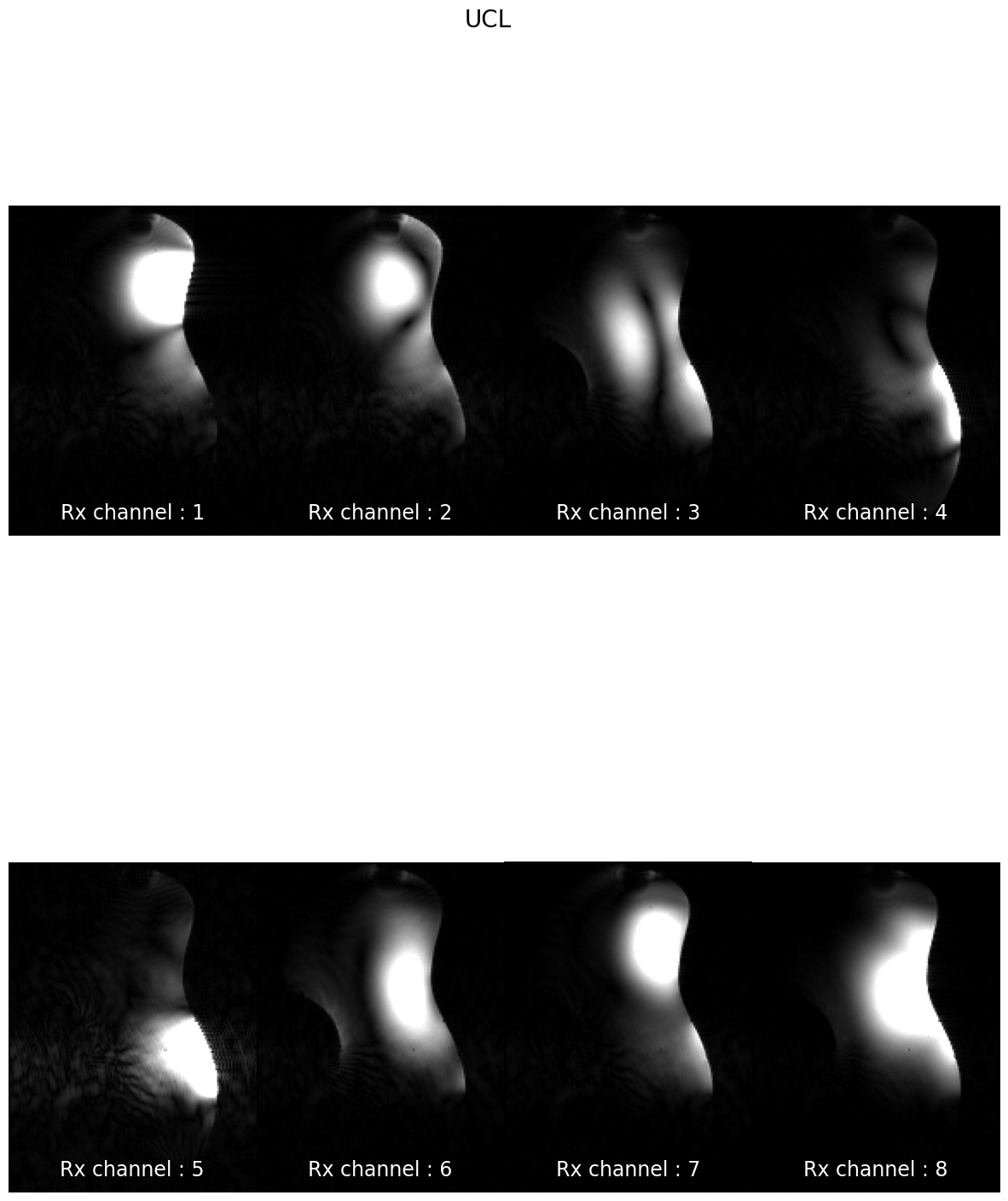
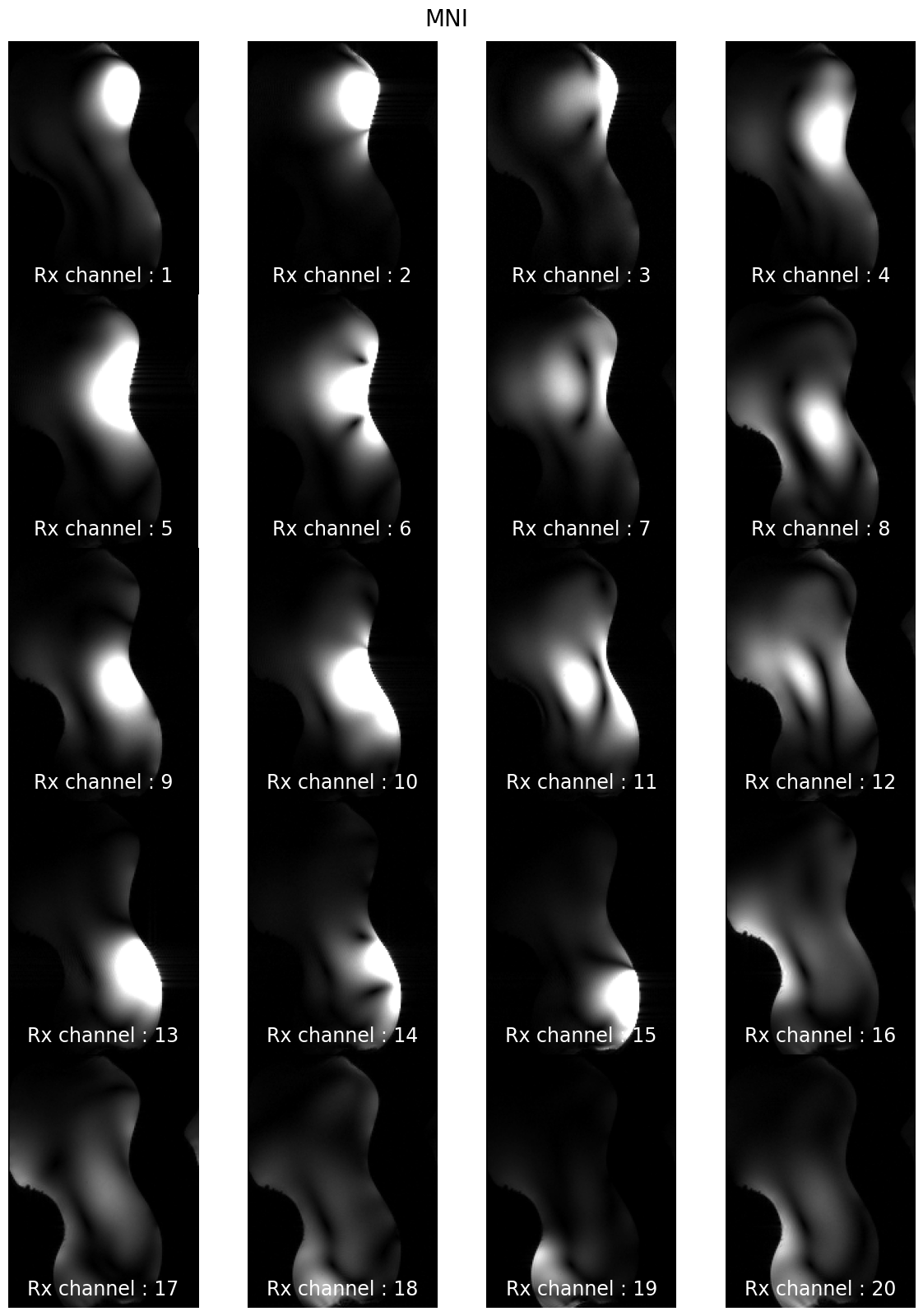
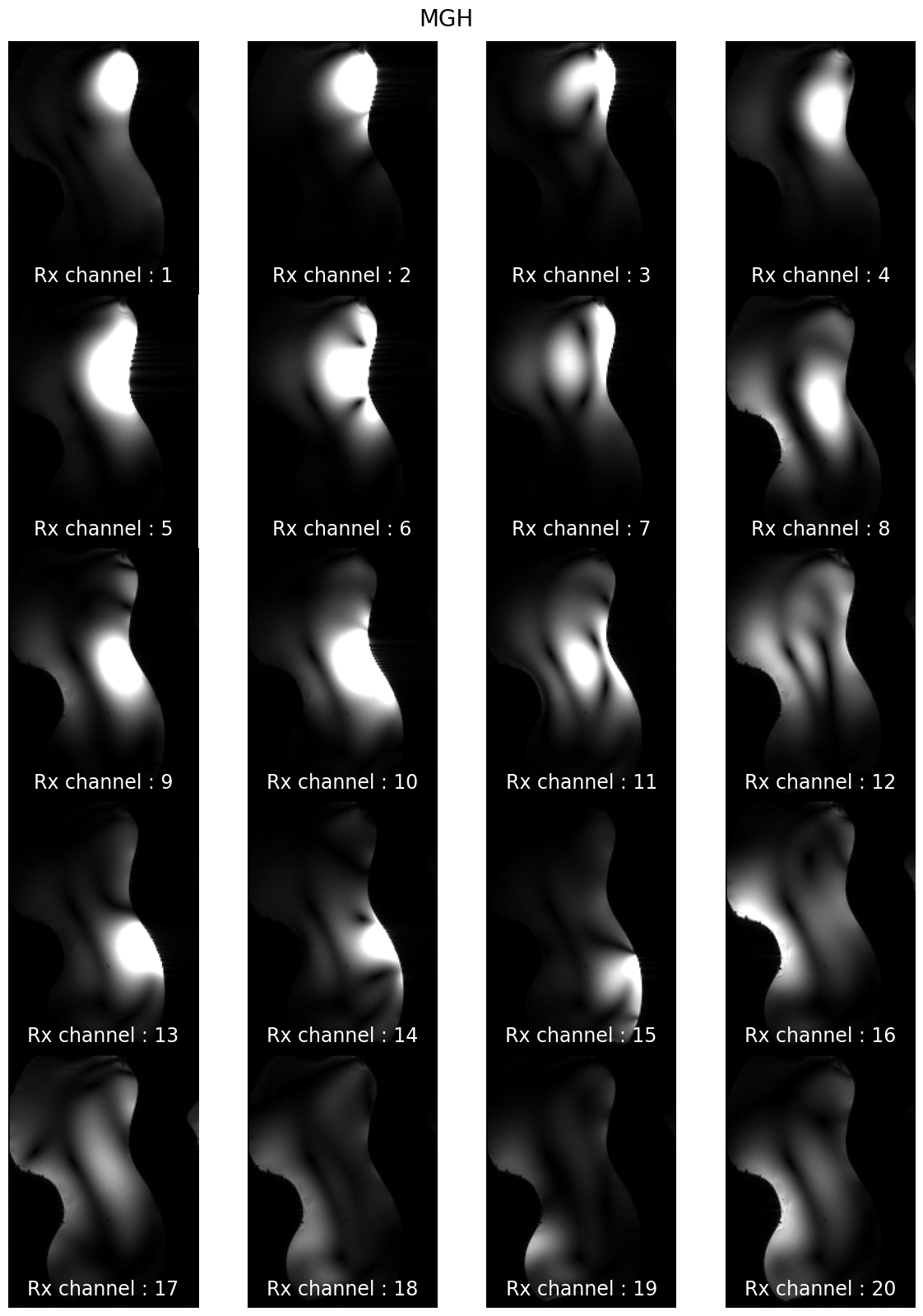
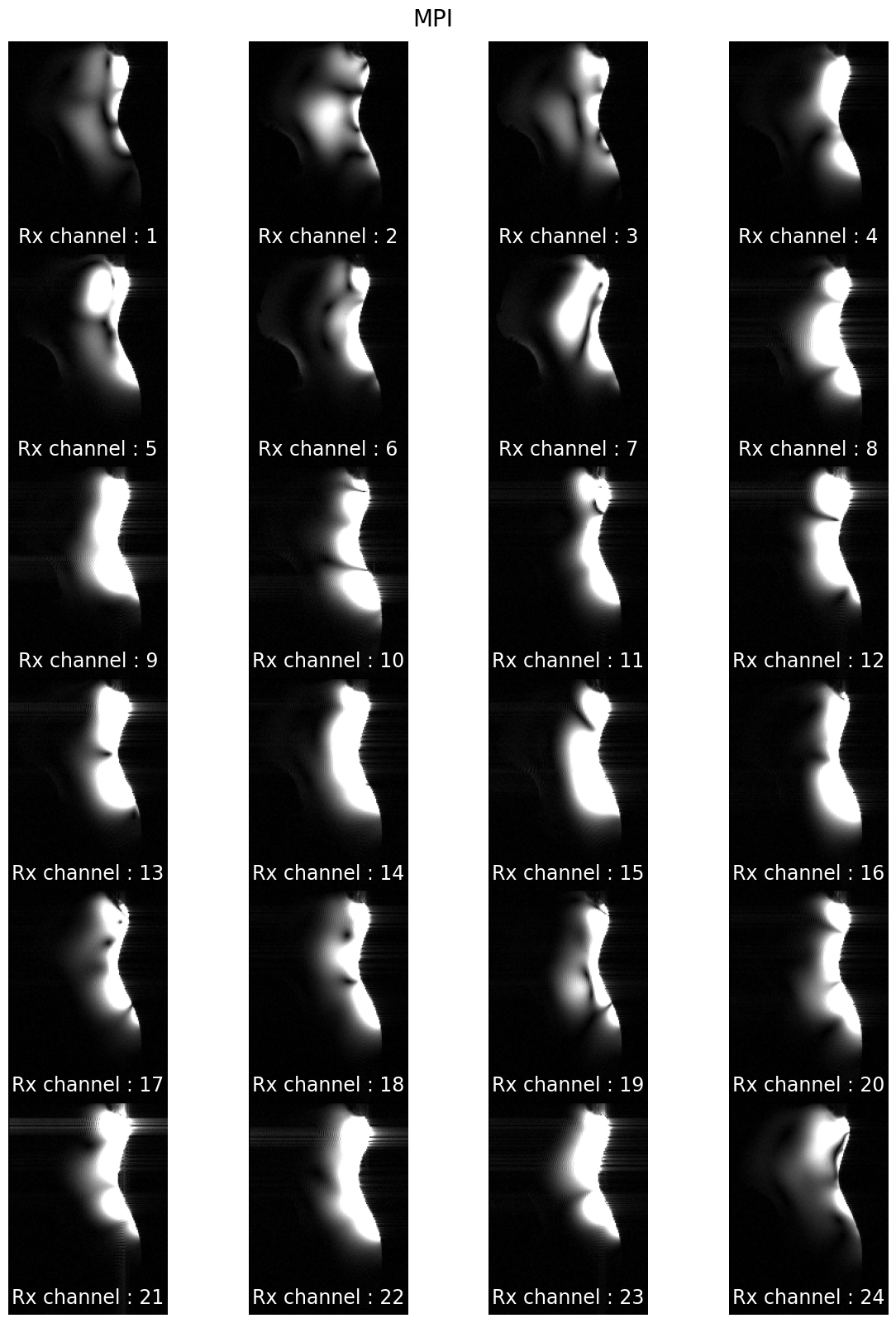
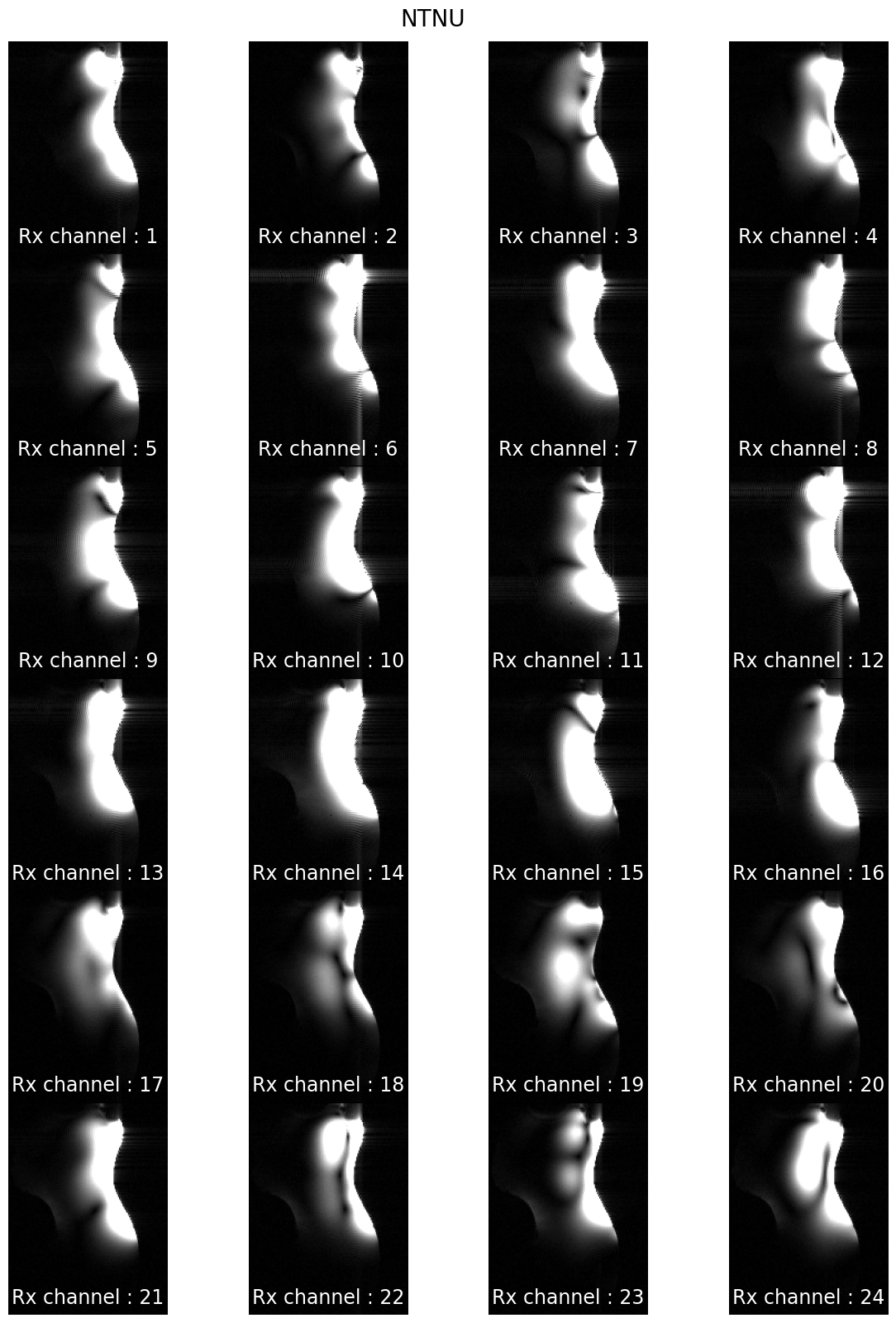
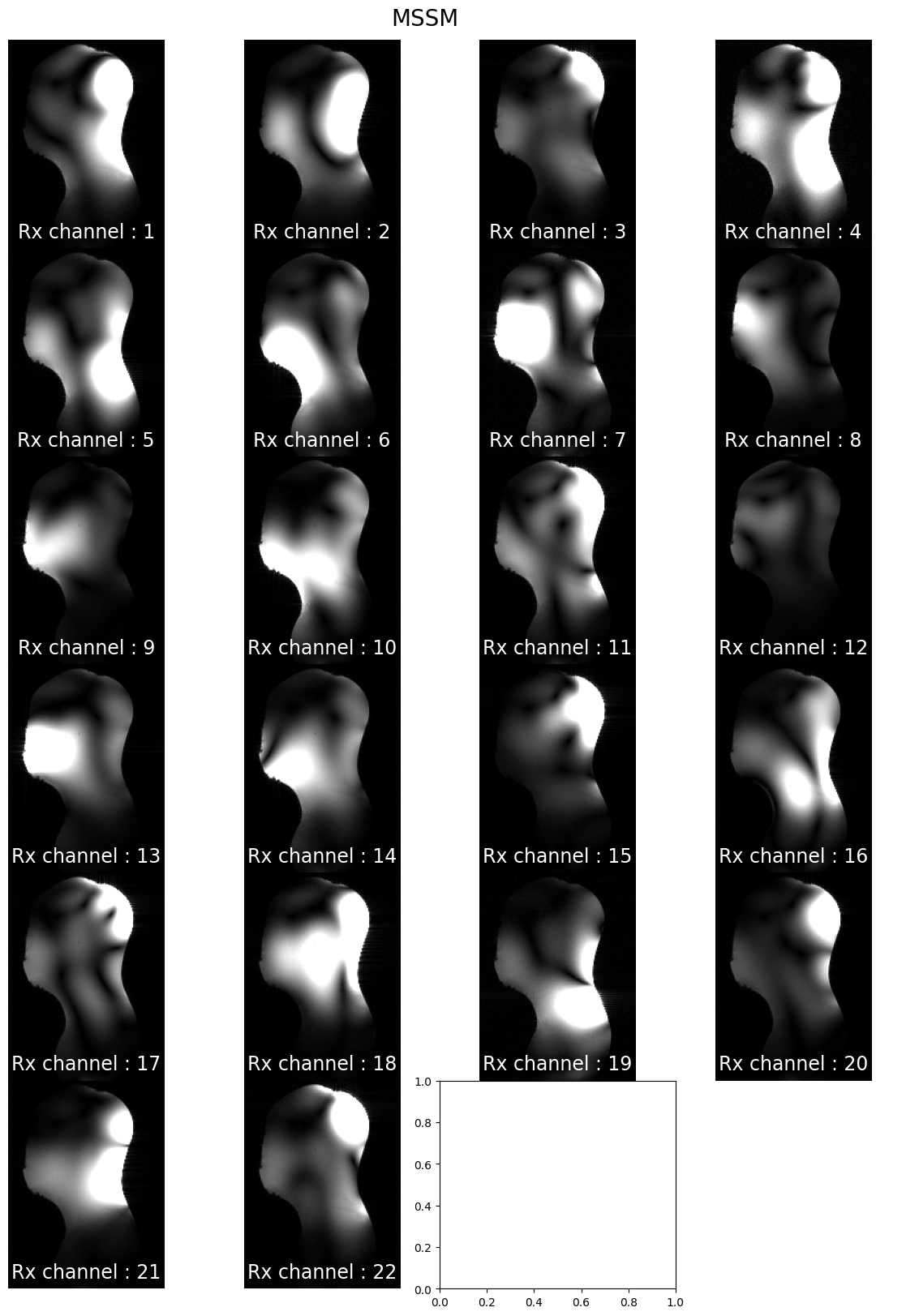
Finished#
# Indicate duration of data processing
end_time = datetime.now()
total_time = (end_time - start_time).total_seconds()
# Convert seconds to a timedelta object
total_time_delta = timedelta(seconds=total_time)
# Format the timedelta object to a string
formatted_time = str(total_time_delta)
# Pad the string representation if less than an hour
formatted_time = formatted_time.rjust(8, '0')
print(f"Total Runtime [hour:min:sec]: {formatted_time}")
Total Runtime [hour:min:sec]: 0:04:50.734726
