7T coil comparison: processing of human data#
Setup environment#
# Necessary imports
from datetime import datetime, timedelta
import os
import json
import glob
import matplotlib.pyplot as plt
import numpy as np
import nibabel as nib
import pandas as pd
from scipy.stats import iqr
import shutil
import zipfile
from scipy.interpolate import interp1d
from scipy.ndimage import uniform_filter1d
from pathlib import Path
%matplotlib inline
start_time = datetime.now()
# Download data from OpenNeuro ⏳
!openneuro-py download --dataset ds005025 --target-dir data-human/
👋 Hello! This is openneuro-py 2024.2.0. Great to see you! 🤗
👉 Please report problems 🤯 and bugs 🪲 at
https://github.com/hoechenberger/openneuro-py/issues
🌍 Preparing to download ds005025 …
📁 Traversing directories for ds005025 : 0 entities [00:00, ? entities/s]
📁 Traversing directories for ds005025 : 5 entities [00:00, 14.15 entities/s]
📁 Traversing directories for ds005025 : 7 entities [00:00, 11.72 entities/s]
📁 Traversing directories for ds005025 : 9 entities [00:00, 8.95 entities/s]
📁 Traversing directories for ds005025 : 11 entities [00:01, 8.97 entities/s]
📁 Traversing directories for ds005025 : 13 entities [00:01, 9.04 entities/s]
📁 Traversing directories for ds005025 : 15 entities [00:01, 10.72 entities/s]
📁 Traversing directories for ds005025 : 17 entities [00:01, 10.17 entities/s]
📁 Traversing directories for ds005025 : 19 entities [00:01, 9.90 entities/s]
📁 Traversing directories for ds005025 : 21 entities [00:01, 11.45 entities/s]
📁 Traversing directories for ds005025 : 23 entities [00:02, 13.07 entities/s]
📁 Traversing directories for ds005025 : 27 entities [00:02, 14.26 entities/s]
📁 Traversing directories for ds005025 : 31 entities [00:02, 18.81 entities/s]
📁 Traversing directories for ds005025 : 34 entities [00:02, 16.93 entities/s]
📁 Traversing directories for ds005025 : 36 entities [00:02, 12.81 entities/s]
📁 Traversing directories for ds005025 : 39 entities [00:03, 14.11 entities/s]
📁 Traversing directories for ds005025 : 42 entities [00:03, 15.52 entities/s]
📁 Traversing directories for ds005025 : 44 entities [00:03, 16.00 entities/s]
📁 Traversing directories for ds005025 : 50 entities [00:03, 22.94 entities/s]
📁 Traversing directories for ds005025 : 53 entities [00:03, 23.99 entities/s]
📁 Traversing directories for ds005025 : 56 entities [00:03, 20.31 entities/s]
📁 Traversing directories for ds005025 : 59 entities [00:03, 21.41 entities/s]
📁 Traversing directories for ds005025 : 104 entities [00:04, 96.57 entities/s]
📁 Traversing directories for ds005025 : 150 entities [00:04, 151.76 entities/s]
📁 Traversing directories for ds005025 : 196 entities [00:04, 192.32 entities/s]
📁 Traversing directories for ds005025 : 266 entities [00:04, 260.36 entities/s]
📁 Traversing directories for ds005025 : 336 entities [00:04, 313.18 entities/s]
📁 Traversing directories for ds005025 : 406 entities [00:04, 346.41 entities/s]
📁 Traversing directories for ds005025 : 476 entities [00:05, 364.57 entities/s]
📁 Traversing directories for ds005025 : 524 entities [00:05, 386.01 entities/s]
📁 Traversing directories for ds005025 : 564 entities [00:05, 386.25 entities/s]
📁 Traversing directories for ds005025 : 612 entities [00:05, 333.54 entities/s]
📁 Traversing directories for ds005025 : 690 entities [00:05, 366.24 entities/s]
📁 Traversing directories for ds005025 : 764 entities [00:05, 400.34 entities/s]
📁 Traversing directories for ds005025 : 842 entities [00:06, 416.44 entities/s]
📁 Traversing directories for ds005025 : 932 entities [00:06, 443.75 entities/s]
📁 Traversing directories for ds005025 : 1030 entities [00:06, 489.25 entities/s]
📁 Traversing directories for ds005025 : 1124 entities [00:06, 502.00 entities/s]
📁 Traversing directories for ds005025 : 1198 entities [00:06, 484.90 entities/s]
📁 Traversing directories for ds005025 : 1254 entities [00:06, 396.98 entities/s]
📁 Traversing directories for ds005025 : 1296 entities [00:07, 396.58 entities/s]
📁 Traversing directories for ds005025 : 1350 entities [00:07, 358.80 entities/s]
📁 Traversing directories for ds005025 : 1396 entities [00:07, 342.49 entities/s]
📁 Traversing directories for ds005025 : 1442 entities [00:07, 324.13 entities/s]
📁 Traversing directories for ds005025 : 1487 entities [00:07, 194.24 entities/s]
📥 Retrieving up to 1487 files (5 concurrent downloads).
.bidsignore: 0%| | 0.00/92.0 [00:00<?, ?B/s]
dataset_description.json: 0%| | 0.00/306 [00:00<?, ?B/s]
sub-CRMBM1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
CHANGES: 0%| | 0.00/1.16k [00:00<?, ?B/s]
participants.tsv: 0%| | 0.00/945 [00:00<?, ?B/s]
sub-CRMBM1_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-CRMBM3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-CRMBM2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-CRMBM3_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1_label-disc.nii.gz: 100%|███| 73.2k/73.2k [00:00<00:00, 572kB/s]
sub-CRMBM3_UNIT1_label-disc.nii.gz: 91%|██▋| 66.6k/73.2k [00:00<00:00, 530kB/s]
sub-MGH1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MGH1_UNIT1_label-disc.nii.gz: 0%| | 0.00/47.1k [00:00<?, ?B/s]
sub-MGH2_UNIT1_label-disc.nii.gz: 0%| | 0.00/47.2k [00:00<?, ?B/s]
sub-MGH2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MGH3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MGH3_UNIT1_label-disc.nii.gz: 0%| | 0.00/47.1k [00:00<?, ?B/s]
sub-MNI1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MNI1_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.1k [00:00<?, ?B/s]
sub-MNI2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MNI1_UNIT1_label-disc.nii.gz: 100%|█████| 73.1k/73.1k [00:00<00:00, 606kB/s]
sub-MNI2_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.1k [00:00<?, ?B/s]
sub-MNI3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MNI3_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-MPI1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MPI1_UNIT1_label-SC_seg.nii.gz: 0%| | 0.00/83.2k [00:00<?, ?B/s]
sub-MNI3_UNIT1_label-disc.nii.gz: 93%|████▋| 68.2k/73.2k [00:00<00:00, 501kB/s]
sub-MPI1_UNIT1_label-SC_seg.json: 0%| | 0.00/292 [00:00<?, ?B/s]
sub-MPI1_UNIT1_label-SC_seg.nii.gz: 100%|███| 83.2k/83.2k [00:00<00:00, 564kB/s]
sub-MPI1_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-MPI2_UNIT1_label-SC_seg.json: 0%| | 0.00/466 [00:00<?, ?B/s]
sub-MPI2_UNIT1_label-SC_seg.json: 100%|████████| 466/466 [00:00<00:00, 3.85kB/s]
sub-MPI2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MPI2_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-MPI3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MPI2_UNIT1_label-SC_seg.nii.gz: 0%| | 0.00/84.3k [00:00<?, ?B/s]
sub-MSSM1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MPI2_UNIT1_label-SC_seg.nii.gz: 99%|██▉| 83.6k/84.3k [00:00<00:00, 606kB/s]
sub-MPI3_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-MSSM1_acq-anat_TB1TFL_label-SC_seg.nii.gz: 0%| | 0.00/25.3k [00:00<?, ?B/s
sub-MSSM1_UNIT1_label-disc.nii.gz: 0%| | 0.00/72.5k [00:00<?, ?B/s]
sub-MSSM2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MSSM2_UNIT1_label-disc.nii.gz: 0%| | 0.00/72.5k [00:00<?, ?B/s]
sub-MSSM2_acq-anat_TB1TFL_label-SC_seg.nii.gz: 0%| | 0.00/25.0k [00:00<?, ?B/s
sub-MSSM3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-MSSM3_UNIT1_label-disc.nii.gz: 0%| | 0.00/72.5k [00:00<?, ?B/s]
sub-NTNU1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-NTNU1_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-MSSM3_acq-anat_TB1TFL_label-SC_seg.nii.gz: 0%| | 0.00/26.7k [00:00<?, ?B/s
sub-NTNU2_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-NTNU2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-NTNU3_UNIT1_label-SC_seg.json: 0%| | 0.00/449 [00:00<?, ?B/s]
sub-NTNU3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-NTNU3_UNIT1_label-SC_seg.nii.gz: 0%| | 0.00/86.1k [00:00<?, ?B/s]
sub-NTNU3_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-NTNU3_acq-anat_TB1TFL_label-SC_seg.json: 0%| | 0.00/449 [00:00<?, ?B/s]
sub-NTNU3_acq-anat_TB1TFL_label-SC_seg.nii.gz: 0%| | 0.00/3.73k [00:00<?, ?B/s
sub-NTNU3_UNIT1_label-SC_seg.nii.gz: 99%|█▉| 85.2k/86.1k [00:00<00:00, 628kB/s]
sub-UCL1_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-UCL2_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-UCL2_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-UCL3_UNIT1_label-disc.json: 0%| | 0.00/198 [00:00<?, ?B/s]
sub-UCL1_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-UCL3_UNIT1_label-disc.nii.gz: 0%| | 0.00/73.2k [00:00<?, ?B/s]
sub-CRMBM1_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-CRMBM1_UNIT1.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-CRMBM1_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-CRMBM1_inv-1_MP2RAGE.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-CRMBM1_T2starw.nii.gz: 8%|▉ | 83.6k/1.06M [00:00<00:01, 608kB/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/15.6M [00:00<?, ?B/s]
sub-CRMBM1_UNIT1.nii.gz: 0%| | 0.00/26.6M [00:00<?, ?B/s]
sub-CRMBM1_T2starw.nii.gz: 33%|███▉ | 356k/1.06M [00:00<00:00, 1.41MB/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/15.6M [00:00<00:24, 652k
sub-CRMBM1_UNIT1.nii.gz: 0%| | 83.5k/26.6M [00:00<00:43, 644kB/s]
sub-CRMBM1_inv-2_MP2RAGE.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.5M [00:00<?, ?B/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 356k/15.6M [00:00<00:10, 1.51M
sub-CRMBM1_UNIT1.nii.gz: 1%|▏ | 373k/26.6M [00:00<00:17, 1.58MB/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 8%| | 1.31M/15.6M [00:00<00:03, 4.86
sub-CRMBM1_UNIT1.nii.gz: 5%|▋ | 1.33M/26.6M [00:00<00:05, 4.89MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.5k/17.5M [00:00<00:30, 607k
sub-CRMBM1_rec-uncombined1_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.01M/15.6M [00:00<00:02, 5.73
sub-CRMBM1_UNIT1.nii.gz: 8%|█ | 2.12M/26.6M [00:00<00:04, 6.09MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 390k/17.5M [00:00<00:11, 1.52M
sub-CRMBM1_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/805k [00:00<?, ?B/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 27%|▎| 4.17M/15.6M [00:00<00:01, 11.4
sub-CRMBM1_UNIT1.nii.gz: 16%|██▏ | 4.37M/26.6M [00:00<00:01, 12.0MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.08M/17.5M [00:00<00:04, 3.66
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 41%|▍| 6.34M/15.6M [00:00<00:00, 15.1
sub-CRMBM1_UNIT1.nii.gz: 24%|███▏ | 6.51M/26.6M [00:00<00:01, 15.3MB/s]
sub-CRMBM1_rec-uncombined1_T2starw.nii.gz: 10%| | 83.5k/805k [00:00<00:01, 649k
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 2.61M/17.5M [00:00<00:01, 7.91
sub-CRMBM1_rec-uncombined2_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_UNIT1.nii.gz: 30%|███▉ | 8.07M/26.6M [00:00<00:01, 15.3MB/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 51%|▌| 8.03M/15.6M [00:00<00:00, 15.1
sub-CRMBM1_rec-uncombined1_T2starw.nii.gz: 29%|▎| 237k/805k [00:00<00:00, 1.12M
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 22%|▏| 3.87M/17.5M [00:00<00:01, 9.65
sub-CRMBM1_UNIT1.nii.gz: 37%|████▊ | 9.76M/26.6M [00:00<00:01, 16.0MB/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 62%|▌| 9.70M/15.6M [00:00<00:00, 15.8
sub-CRMBM1_rec-uncombined1_T2starw.nii.gz: 93%|▉| 747k/805k [00:00<00:00, 2.86M
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 30%|▎| 5.17M/17.5M [00:00<00:01, 10.9
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 72%|▋| 11.2M/15.6M [00:00<00:00, 15.5
sub-CRMBM1_UNIT1.nii.gz: 43%|█████▌ | 11.3M/26.6M [00:00<00:01, 15.6MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 40%|▍| 7.09M/17.5M [00:00<00:00, 13.8
sub-CRMBM1_rec-uncombined3_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 82%|▊| 12.8M/15.6M [00:01<00:00, 15.4
sub-CRMBM1_UNIT1.nii.gz: 48%|██████▎ | 12.8M/26.6M [00:01<00:00, 15.5MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 48%|▍| 8.48M/17.5M [00:00<00:00, 14.0
sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz: 95%|▉| 14.8M/15.6M [00:01<00:00, 17.1
sub-CRMBM1_UNIT1.nii.gz: 56%|███████▏ | 14.8M/26.6M [00:01<00:00, 17.1MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 60%|▌| 10.5M/17.5M [00:00<00:00, 16.3
sub-CRMBM1_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/826k [00:00<?, ?B/s]
sub-CRMBM1_UNIT1.nii.gz: 62%|████████ | 16.5M/26.6M [00:01<00:00, 17.2MB/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 71%|▋| 12.5M/17.5M [00:01<00:00, 17.5
sub-CRMBM1_rec-uncombined2_T2starw.nii.gz: 10%| | 83.6k/826k [00:00<00:01, 727k
sub-CRMBM1_UNIT1.nii.gz: 71%|█████████▏ | 18.8M/26.6M [00:01<00:00, 19.3MB/s]
sub-CRMBM1_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/886k [00:00<?, ?B/s]
sub-CRMBM1_inv-2_part-mag_MP2RAGE.nii.gz: 86%|▊| 15.1M/17.5M [00:01<00:00, 20.4
sub-CRMBM1_UNIT1.nii.gz: 78%|██████████▏ | 20.8M/26.6M [00:01<00:00, 19.8MB/s]
sub-CRMBM1_rec-uncombined2_T2starw.nii.gz: 41%|▍| 339k/826k [00:00<00:00, 1.50M
sub-CRMBM1_rec-uncombined3_T2starw.nii.gz: 9%| | 77.0k/886k [00:00<00:01, 591k
sub-CRMBM1_rec-uncombined4_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_UNIT1.nii.gz: 85%|███████████ | 22.7M/26.6M [00:01<00:00, 17.2MB/s]
sub-CRMBM1_rec-uncombined3_T2starw.nii.gz: 41%|▍| 366k/886k [00:00<00:00, 1.58M
sub-CRMBM1_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/841k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined5_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/777k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined5_T2starw.nii.gz: 10%| | 83.6k/841k [00:00<00:01, 655k
sub-CRMBM1_rec-uncombined6_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/845k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined5_T2starw.nii.gz: 42%|▍| 356k/841k [00:00<00:00, 1.51M
sub-CRMBM1_rec-uncombined4_T2starw.nii.gz: 11%| | 83.6k/777k [00:00<00:01, 529k
sub-CRMBM1_rec-uncombined7_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined6_T2starw.nii.gz: 10%| | 83.5k/845k [00:00<00:01, 611k
sub-CRMBM1_rec-uncombined4_T2starw.nii.gz: 46%|▍| 356k/777k [00:00<00:00, 1.45M
sub-CRMBM1_rec-uncombined6_T2starw.nii.gz: 48%|▍| 407k/845k [00:00<00:00, 1.64M
sub-CRMBM1_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/804k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined8_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined7_T2starw.nii.gz: 6%| | 52.2k/804k [00:00<00:01, 522k
sub-CRMBM1_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/797k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined7_T2starw.nii.gz: 20%|▏| 162k/804k [00:00<00:00, 846kB
sub-CRMBM1_acq-anat_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined8_T2starw.nii.gz: 10%| | 83.6k/797k [00:00<00:01, 651k
sub-CRMBM1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM1_rec-uncombined7_T2starw.nii.gz: 90%|▉| 723k/804k [00:00<00:00, 2.41M
sub-CRMBM1_rec-uncombined8_T2starw.nii.gz: 45%|▍| 356k/797k [00:00<00:00, 1.51M
sub-CRMBM1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/854k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.21M [00:00<?, ?B/s]
sub-CRMBM1_acq-anat_TB1TFL.nii.gz: 10%|▍ | 85.2k/854k [00:00<00:01, 629kB/s]
sub-CRMBM1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 77.0k/4.21M [00:00<00:07, 602k
sub-CRMBM1_acq-anat_TB1TFL.nii.gz: 44%|██▏ | 375k/854k [00:00<00:00, 1.51MB/s]
sub-CRMBM1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.69M [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 383k/4.21M [00:00<00:02, 1.65M
sub-CRMBM1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.69M [00:00<00:04,
sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz: 36%|▎| 1.54M/4.21M [00:00<00:00, 5.13
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaSagSmall_GFactor.nii.gz: 12%| | 339k/2.69M [00:00<00:01, 1
sub-CRMBM1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 1%| | 67.6k/11.7M [00:00<00:18, 649kB
sub-CRMBM1_acq-coilQaSagSmall_GFactor.nii.gz: 27%|▎| 748k/2.69M [00:00<00:00, 2
sub-CRMBM1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.7k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 1%| | 169k/11.7M [00:00<00:14, 857kB/
sub-CRMBM1_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM1_acq-famp-0.66_TB1DREAM.nii.gz: 92%|▉| 85.2k/92.7k [00:00<00:00, 566k
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 6%| | 730k/11.7M [00:00<00:04, 2.50MB
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 23%|▏| 2.75M/11.7M [00:00<00:01, 8.96M
sub-CRMBM1_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/93.4k [00:00<?, ?B/s]
sub-CRMBM1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/93.3k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 47%|▍| 5.57M/11.7M [00:00<00:00, 15.6M
sub-CRMBM1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-CRMBM1_acq-famp-1.5_TB1DREAM.nii.gz: 89%|▉| 83.5k/93.4k [00:00<00:00, 518kB
sub-CRMBM1_acq-famp_TB1TFL.json: 0%| | 0.00/2.70k [00:00<?, ?B/s]
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 68%|▋| 7.96M/11.7M [00:00<00:00, 18.1M
sub-CRMBM1_acq-famp_TB1DREAM.nii.gz: 90%|█▊| 83.6k/93.3k [00:00<00:00, 478kB/s]
sub-CRMBM1_acq-coilQaTra_GFactor.nii.gz: 92%|▉| 10.8M/11.7M [00:00<00:00, 21.7M
sub-CRMBM1_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-CRMBM1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/949k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/86.5k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.6k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv-0.66_TB1DREAM.nii.gz: 97%|▉| 83.6k/86.5k [00:00<00:00, 707k
sub-CRMBM1_acq-famp_TB1TFL.nii.gz: 9%|▍ | 85.2k/949k [00:00<00:01, 583kB/s]
sub-CRMBM1_acq-refv-1.5_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.6k [00:00<00:00, 613kB
sub-CRMBM2_T2starw.json: 0%| | 0.00/2.47k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-CRMBM1_acq-famp_TB1TFL.nii.gz: 36%|█▊ | 340k/949k [00:00<00:00, 1.38MB/s]
sub-CRMBM2_T2starw.nii.gz: 0%| | 0.00/990k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/90.2k [00:00<?, ?B/s]
sub-CRMBM2_T2starw.nii.gz: 8%|█ | 78.0k/990k [00:00<00:01, 608kB/s]
sub-CRMBM2_inv-1_MP2RAGE.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 0%| | 0.00/26.8M [00:00<?, ?B/s]
sub-CRMBM2_T2starw.nii.gz: 37%|████▊ | 366k/990k [00:00<00:00, 1.57MB/s]
sub-CRMBM1_acq-refv_TB1DREAM.nii.gz: 93%|█▊| 83.6k/90.2k [00:00<00:00, 615kB/s]
sub-CRMBM2_UNIT1.nii.gz: 0%| | 83.6k/26.8M [00:00<00:47, 594kB/s]
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/13.7M [00:00<?, ?B/s]
sub-CRMBM2_inv-2_MP2RAGE.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 1%|▏ | 356k/26.8M [00:00<00:17, 1.54MB/s]
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/13.7M [00:00<00:22, 643k
sub-CRMBM2_rec-uncombined1_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 5%|▋ | 1.29M/26.8M [00:00<00:05, 4.56MB/s]
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 339k/13.7M [00:00<00:09, 1.43M
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/15.7M [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 12%|█▌ | 3.34M/26.8M [00:00<00:02, 10.4MB/s]
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 9%| | 1.19M/13.7M [00:00<00:03, 4.38
sub-CRMBM2_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/744k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 28%|███▌ | 7.47M/26.8M [00:00<00:00, 21.2MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/15.7M [00:00<00:25, 650k
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 20%|▏| 2.80M/13.7M [00:00<00:01, 8.82
sub-CRMBM2_UNIT1.nii.gz: 39%|█████ | 10.4M/26.8M [00:00<00:00, 24.3MB/s]
sub-CRMBM2_rec-uncombined1_T2starw.nii.gz: 11%| | 83.6k/744k [00:00<00:01, 614k
sub-CRMBM2_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/753k [00:00<?, ?B/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 339k/15.7M [00:00<00:11, 1.42M
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 37%|▎| 5.10M/13.7M [00:00<00:00, 13.9
sub-CRMBM2_UNIT1.nii.gz: 48%|██████▏ | 12.9M/26.8M [00:00<00:00, 24.2MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 7%| | 1.08M/15.7M [00:00<00:03, 3.91
sub-CRMBM2_rec-uncombined2_T2starw.nii.gz: 11%| | 83.6k/753k [00:00<00:01, 615k
sub-CRMBM2_rec-uncombined1_T2starw.nii.gz: 43%|▍| 322k/744k [00:00<00:00, 1.25M
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 51%|▌| 7.02M/13.7M [00:00<00:00, 15.9
sub-CRMBM2_UNIT1.nii.gz: 57%|███████▍ | 15.3M/26.8M [00:00<00:00, 22.1MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.03M/15.7M [00:00<00:02, 6.04
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 63%|▋| 8.62M/13.7M [00:00<00:00, 14.9
sub-CRMBM2_rec-uncombined2_T2starw.nii.gz: 47%|▍| 356k/753k [00:00<00:00, 1.44M
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 21%|▏| 3.29M/15.7M [00:00<00:01, 8.45
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 73%|▋| 10.1M/13.7M [00:00<00:00, 14.5
sub-CRMBM2_UNIT1.nii.gz: 65%|████████▍ | 17.5M/26.8M [00:01<00:00, 18.1MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 30%|▎| 4.75M/15.7M [00:00<00:01, 10.6
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 87%|▊| 12.0M/13.7M [00:01<00:00, 16.0
sub-CRMBM2_UNIT1.nii.gz: 72%|█████████▍ | 19.4M/26.8M [00:01<00:00, 18.5MB/s]
sub-CRMBM2_rec-uncombined2_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 41%|▍| 6.50M/15.7M [00:00<00:00, 12.5
sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz: 99%|▉| 13.6M/13.7M [00:01<00:00, 15.9
sub-CRMBM2_UNIT1.nii.gz: 79%|██████████▎ | 21.2M/26.8M [00:01<00:00, 16.9MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 51%|▌| 8.06M/15.7M [00:00<00:00, 13.7
sub-CRMBM2_UNIT1.nii.gz: 89%|███████████▌ | 23.8M/26.8M [00:01<00:00, 19.4MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 68%|▋| 10.7M/15.7M [00:00<00:00, 17.8
sub-CRMBM2_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/815k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined3_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_UNIT1.nii.gz: 97%|████████████▋| 26.1M/26.8M [00:01<00:00, 20.3MB/s]
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 81%|▊| 12.7M/15.7M [00:01<00:00, 18.8
sub-CRMBM2_rec-uncombined4_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined3_T2starw.nii.gz: 10%| | 83.6k/815k [00:00<00:01, 515k
sub-CRMBM2_inv-2_part-mag_MP2RAGE.nii.gz: 94%|▉| 14.8M/15.7M [00:01<00:00, 19.7
sub-CRMBM2_rec-uncombined3_T2starw.nii.gz: 44%|▍| 356k/815k [00:00<00:00, 1.47M
sub-CRMBM2_rec-uncombined5_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/723k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/771k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined6_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined4_T2starw.nii.gz: 12%| | 85.2k/723k [00:00<00:00, 666k
sub-CRMBM2_rec-uncombined5_T2starw.nii.gz: 11%| | 85.2k/771k [00:00<00:01, 639k
sub-CRMBM2_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/753k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined4_T2starw.nii.gz: 49%|▍| 357k/723k [00:00<00:00, 1.52M
sub-CRMBM2_rec-uncombined8_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined5_T2starw.nii.gz: 40%|▍| 306k/771k [00:00<00:00, 1.15M
sub-CRMBM2_rec-uncombined6_T2starw.nii.gz: 11%| | 83.6k/753k [00:00<00:01, 655k
sub-CRMBM2_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/731k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined6_T2starw.nii.gz: 52%|▌| 390k/753k [00:00<00:00, 1.68M
sub-CRMBM2_rec-uncombined8_T2starw.nii.gz: 11%| | 78.0k/731k [00:00<00:01, 607k
sub-CRMBM2_rec-uncombined7_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/700k [00:00<?, ?B/s]
sub-CRMBM2_acq-anat_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined8_T2starw.nii.gz: 50%|▌| 366k/731k [00:00<00:00, 1.50M
sub-CRMBM2_rec-uncombined7_T2starw.nii.gz: 12%| | 83.6k/700k [00:00<00:00, 655k
sub-CRMBM2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-CRMBM2_rec-uncombined7_T2starw.nii.gz: 51%|▌| 356k/700k [00:00<00:00, 1.52M
sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.02M [00:00<?, ?B/s]
sub-CRMBM2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/802k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.24k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 77.0k/4.02M [00:00<00:06, 674k
sub-CRMBM2_acq-anat_TB1TFL.nii.gz: 10%|▌ | 83.5k/802k [00:00<00:01, 646kB/s]
sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 349k/4.02M [00:00<00:02, 1.50M
sub-CRMBM2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-CRMBM2_acq-anat_TB1TFL.nii.gz: 29%|█▍ | 237k/802k [00:00<00:00, 1.12MB/s]
sub-CRMBM2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.69M [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz: 27%|▎| 1.09M/4.02M [00:00<00:00, 3.98
sub-CRMBM2_acq-anat_TB1TFL.nii.gz: 93%|████▋| 748k/802k [00:00<00:00, 2.87MB/s]
sub-CRMBM2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 94.0k/2.69M [00:00<00:04,
sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz: 47%|▍| 1.87M/4.02M [00:00<00:00, 5.49
sub-CRMBM2_acq-coilQaSagSmall_GFactor.nii.gz: 15%|▏| 400k/2.69M [00:00<00:01, 1
sub-CRMBM2_acq-coilQaSagSmall_GFactor.nii.gz: 40%|▍| 1.09M/2.69M [00:00<00:00,
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-CRMBM2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.5k [00:00<?, ?B/s]
sub-CRMBM2_acq-famp-0.66_TB1DREAM.nii.gz: 83%|▊| 77.0k/92.5k [00:00<00:00, 642k
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/11.7M [00:00<00:25, 480kB
sub-CRMBM2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.7k [00:00<?, ?B/s]
sub-CRMBM2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 3%| | 356k/11.7M [00:00<00:08, 1.39MB
sub-CRMBM2_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 85.2k/91.7k [00:00<00:00, 663kB
sub-CRMBM2_acq-famp_TB1TFL.json: 0%| | 0.00/2.74k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 11%| | 1.26M/11.7M [00:00<00:02, 4.24M
sub-CRMBM2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 25%|▎| 2.94M/11.7M [00:00<00:01, 8.72M
sub-CRMBM2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 63%|▋| 7.34M/11.7M [00:00<00:00, 19.7M
sub-CRMBM2_acq-famp_TB1DREAM.nii.gz: 93%|█▊| 85.2k/92.1k [00:00<00:00, 590kB/s]
sub-CRMBM2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/929k [00:00<?, ?B/s]
sub-CRMBM2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/80.0k [00:00<?, ?B/s]
sub-CRMBM2_acq-coilQaTra_GFactor.nii.gz: 96%|▉| 11.3M/11.7M [00:00<00:00, 26.3M
sub-CRMBM2_acq-famp_TB1TFL.nii.gz: 9%|▍ | 83.6k/929k [00:00<00:01, 746kB/s]
sub-CRMBM2_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 80.0k/80.0k [00:00<00:00, 592k
sub-CRMBM2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM2_acq-famp_TB1TFL.nii.gz: 38%|█▉ | 356k/929k [00:00<00:00, 1.59MB/s]
sub-CRMBM2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/84.4k [00:00<?, ?B/s]
sub-CRMBM2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM2_acq-refv-1.5_TB1DREAM.nii.gz: 100%|█| 84.4k/84.4k [00:00<00:00, 701kB
sub-CRMBM3_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-CRMBM2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/83.1k [00:00<?, ?B/s]
sub-CRMBM3_UNIT1.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-CRMBM2_acq-refv_TB1DREAM.nii.gz: 100%|██| 83.1k/83.1k [00:00<00:00, 552kB/s]
sub-CRMBM3_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-CRMBM3_UNIT1.nii.gz: 0%| | 0.00/26.7M [00:00<?, ?B/s]
sub-CRMBM3_inv-2_MP2RAGE.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-CRMBM3_T2starw.nii.gz: 8%|▉ | 83.6k/1.03M [00:00<00:01, 712kB/s]
sub-CRMBM3_UNIT1.nii.gz: 0%| | 85.2k/26.7M [00:00<00:44, 627kB/s]
sub-CRMBM3_T2starw.nii.gz: 34%|████ | 356k/1.03M [00:00<00:00, 1.53MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/15.1M [00:00<?, ?B/s]
sub-CRMBM3_UNIT1.nii.gz: 1%|▏ | 357k/26.7M [00:00<00:19, 1.41MB/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.3M [00:00<?, ?B/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.5k/15.1M [00:00<00:24, 648k
sub-CRMBM3_inv-1_MP2RAGE.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-CRMBM3_UNIT1.nii.gz: 5%|▋ | 1.45M/26.7M [00:00<00:06, 4.38MB/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/17.3M [00:00<00:26, 680k
sub-CRMBM3_UNIT1.nii.gz: 18%|██▎ | 4.86M/26.7M [00:00<00:01, 14.1MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 3%| | 407k/15.1M [00:00<00:08, 1.73M
sub-CRMBM3_rec-uncombined1_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 356k/17.3M [00:00<00:11, 1.48M
sub-CRMBM3_UNIT1.nii.gz: 26%|███▎ | 6.91M/26.7M [00:00<00:01, 16.4MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 9%| | 1.33M/15.1M [00:00<00:02, 4.83
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.11M/17.3M [00:00<00:04, 3.99
sub-CRMBM3_UNIT1.nii.gz: 34%|████▍ | 9.06M/26.7M [00:00<00:01, 18.3MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 17%|▏| 2.55M/15.1M [00:00<00:01, 7.67
sub-CRMBM3_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/788k [00:00<?, ?B/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.27M/17.3M [00:00<00:02, 6.85
sub-CRMBM3_UNIT1.nii.gz: 41%|█████▎ | 11.0M/26.7M [00:00<00:00, 18.6MB/s]
sub-CRMBM3_rec-uncombined3_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 29%|▎| 4.43M/15.1M [00:00<00:00, 11.6
sub-CRMBM3_rec-uncombined1_T2starw.nii.gz: 11%| | 83.5k/788k [00:00<00:01, 642k
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 21%|▏| 3.58M/17.3M [00:00<00:01, 9.16
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 38%|▍| 5.82M/15.1M [00:00<00:00, 12.6
sub-CRMBM3_UNIT1.nii.gz: 48%|██████▎ | 12.8M/26.7M [00:00<00:00, 17.4MB/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 32%|▎| 5.47M/17.3M [00:00<00:00, 12.6
sub-CRMBM3_rec-uncombined1_T2starw.nii.gz: 52%|▌| 407k/788k [00:00<00:00, 1.72M
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 49%|▍| 7.44M/15.1M [00:00<00:00, 14.0
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 39%|▍| 6.72M/17.3M [00:00<00:00, 12.6
sub-CRMBM3_UNIT1.nii.gz: 55%|███████ | 14.6M/26.7M [00:01<00:00, 16.3MB/s]
sub-CRMBM3_rec-uncombined2_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 58%|▌| 8.82M/15.1M [00:00<00:00, 13.0
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 47%|▍| 8.17M/17.3M [00:00<00:00, 13.4
sub-CRMBM3_UNIT1.nii.gz: 61%|███████▉ | 16.2M/26.7M [00:01<00:00, 16.2MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 71%|▋| 10.8M/15.1M [00:00<00:00, 15.2
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 59%|▌| 10.2M/17.3M [00:00<00:00, 15.7
sub-CRMBM3_UNIT1.nii.gz: 68%|████████▊ | 18.2M/26.7M [00:01<00:00, 17.3MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 84%|▊| 12.7M/15.1M [00:01<00:00, 16.7
sub-CRMBM3_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/798k [00:00<?, ?B/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 70%|▋| 12.0M/17.3M [00:01<00:00, 16.8
sub-CRMBM3_UNIT1.nii.gz: 75%|█████████▊ | 20.1M/26.7M [00:01<00:00, 18.1MB/s]
sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz: 97%|▉| 14.7M/15.1M [00:01<00:00, 17.7
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 81%|▊| 13.9M/17.3M [00:01<00:00, 16.7
sub-CRMBM3_UNIT1.nii.gz: 82%|██████████▋ | 21.9M/26.7M [00:01<00:00, 17.4MB/s]
sub-CRMBM3_rec-uncombined2_T2starw.nii.gz: 10%| | 83.6k/798k [00:00<00:01, 583k
sub-CRMBM3_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/861k [00:00<?, ?B/s]
sub-CRMBM3_inv-2_part-mag_MP2RAGE.nii.gz: 96%|▉| 16.5M/17.3M [00:01<00:00, 19.6
sub-CRMBM3_UNIT1.nii.gz: 92%|███████████▉ | 24.5M/26.7M [00:01<00:00, 20.2MB/s]
sub-CRMBM3_rec-uncombined2_T2starw.nii.gz: 49%|▍| 390k/798k [00:00<00:00, 1.68M
sub-CRMBM3_rec-uncombined3_T2starw.nii.gz: 8%| | 66.5k/861k [00:00<00:01, 484k
sub-CRMBM3_UNIT1.nii.gz: 99%|████████████▉| 26.5M/26.7M [00:01<00:00, 19.6MB/s]
sub-CRMBM3_rec-uncombined3_T2starw.nii.gz: 31%|▎| 271k/861k [00:00<00:00, 1.08M
sub-CRMBM3_rec-uncombined4_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/758k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined5_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/817k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined6_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined4_T2starw.nii.gz: 11%| | 83.6k/758k [00:00<00:01, 600k
sub-CRMBM3_rec-uncombined5_T2starw.nii.gz: 10%| | 85.2k/817k [00:00<00:00, 791k
sub-CRMBM3_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/835k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined5_T2starw.nii.gz: 50%|▍| 408k/817k [00:00<00:00, 1.86M
sub-CRMBM3_rec-uncombined4_T2starw.nii.gz: 51%|▌| 390k/758k [00:00<00:00, 1.56M
sub-CRMBM3_rec-uncombined6_T2starw.nii.gz: 10%| | 83.6k/835k [00:00<00:01, 578k
sub-CRMBM3_rec-uncombined7_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined8_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined6_T2starw.nii.gz: 47%|▍| 390k/835k [00:00<00:00, 1.70M
sub-CRMBM3_acq-anat_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/773k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/775k [00:00<?, ?B/s]
sub-CRMBM3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/871k [00:00<?, ?B/s]
sub-CRMBM3_rec-uncombined8_T2starw.nii.gz: 11%| | 85.2k/773k [00:00<00:01, 658k
sub-CRMBM3_rec-uncombined7_T2starw.nii.gz: 11%| | 85.2k/775k [00:00<00:01, 666k
sub-CRMBM3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM3_acq-anat_TB1TFL.nii.gz: 9%|▍ | 77.0k/871k [00:00<00:01, 564kB/s]
sub-CRMBM3_rec-uncombined8_T2starw.nii.gz: 46%|▍| 357k/773k [00:00<00:00, 1.52M
sub-CRMBM3_rec-uncombined7_T2starw.nii.gz: 53%|▌| 408k/775k [00:00<00:00, 1.75M
sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.11M [00:00<?, ?B/s]
sub-CRMBM3_acq-anat_TB1TFL.nii.gz: 44%|██▏ | 383k/871k [00:00<00:00, 1.55MB/s]
sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 85.2k/4.11M [00:00<00:06, 702k
sub-CRMBM3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 357k/4.11M [00:00<00:02, 1.50M
sub-CRMBM3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.69M [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz: 36%|▎| 1.46M/4.11M [00:00<00:00, 4.68
sub-CRMBM3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz: 87%|▊| 3.56M/4.11M [00:00<00:00, 9.99
sub-CRMBM3_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.69M [00:00<00:05,
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaSagSmall_GFactor.nii.gz: 12%| | 339k/2.69M [00:00<00:01, 1
sub-CRMBM3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.4k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 1%| | 94.0k/11.7M [00:00<00:17, 715kB
sub-CRMBM3_acq-coilQaSagSmall_GFactor.nii.gz: 52%|▌| 1.41M/2.69M [00:00<00:00,
sub-CRMBM3_acq-famp-0.66_TB1DREAM.nii.gz: 90%|▉| 83.5k/92.4k [00:00<00:00, 605k
sub-CRMBM3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 3%| | 400k/11.7M [00:00<00:07, 1.61MB
sub-CRMBM3_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/93.0k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 8%| | 995k/11.7M [00:00<00:03, 3.29MB
sub-CRMBM3_acq-famp-1.5_TB1DREAM.nii.gz: 83%|▊| 77.0k/93.0k [00:00<00:00, 602kB
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 24%|▏| 2.81M/11.7M [00:00<00:01, 8.05M
sub-CRMBM3_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 60%|▌| 7.09M/11.7M [00:00<00:00, 19.5M
sub-CRMBM3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-CRMBM3_acq-famp_TB1TFL.json: 0%| | 0.00/2.70k [00:00<?, ?B/s]
sub-CRMBM3_acq-coilQaTra_GFactor.nii.gz: 92%|▉| 10.8M/11.7M [00:00<00:00, 25.4M
sub-CRMBM3_acq-famp_TB1DREAM.nii.gz: 91%|█▊| 83.6k/92.1k [00:00<00:00, 618kB/s]
sub-CRMBM3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/84.0k [00:00<?, ?B/s]
sub-CRMBM3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/954k [00:00<?, ?B/s]
sub-CRMBM3_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-CRMBM3_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 84.0k/84.0k [00:00<00:00, 648k
sub-CRMBM3_acq-famp_TB1TFL.nii.gz: 9%|▍ | 83.6k/954k [00:00<00:01, 556kB/s]
sub-CRMBM3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-CRMBM3_acq-famp_TB1TFL.nii.gz: 37%|█▊ | 356k/954k [00:00<00:00, 1.54MB/s]
sub-CRMBM3_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.2k [00:00<?, ?B/s]
sub-CRMBM3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-CRMBM3_acq-refv-1.5_TB1DREAM.nii.gz: 92%|▉| 83.6k/91.2k [00:00<00:00, 613kB
sub-CRMBM3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/86.1k [00:00<?, ?B/s]
sub-MGH1_T2starw.nii.gz: 0%| | 0.00/1.27M [00:00<?, ?B/s]
sub-MGH1_T2starw.json: 0%| | 0.00/2.42k [00:00<?, ?B/s]
sub-CRMBM3_acq-refv_TB1DREAM.nii.gz: 97%|█▉| 83.6k/86.1k [00:00<00:00, 616kB/s]
sub-MGH1_T2starw.nii.gz: 5%|▋ | 68.2k/1.27M [00:00<00:02, 623kB/s]
sub-MGH1_UNIT1.json: 0%| | 0.00/1.98k [00:00<?, ?B/s]
sub-MGH1_inv-2_MP2RAGE.json: 0%| | 0.00/1.99k [00:00<?, ?B/s]
sub-MGH1_T2starw.nii.gz: 16%|██▎ | 205k/1.27M [00:00<00:01, 971kB/s]
sub-MGH1_T2starw.nii.gz: 55%|███████▋ | 714k/1.27M [00:00<00:00, 2.52MB/s]
sub-MGH1_inv-1_MP2RAGE.json: 0%| | 0.00/1.99k [00:00<?, ?B/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/7.98M [00:00<?, ?B/s]
sub-MGH1_UNIT1.nii.gz: 0%| | 0.00/17.0M [00:00<?, ?B/s]
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/9.54M [00:00<?, ?B/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/7.98M [00:00<00:12, 655kB/
sub-MGH1_UNIT1.nii.gz: 0%| | 83.6k/17.0M [00:00<00:29, 612kB/s]
sub-MGH1_rec-uncombined02_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 3%| | 272k/7.98M [00:00<00:06, 1.32MB/
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/9.54M [00:00<00:17, 569kB/
sub-MGH1_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/666k [00:00<?, ?B/s]
sub-MGH1_UNIT1.nii.gz: 2%|▎ | 356k/17.0M [00:00<00:12, 1.42MB/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 10%| | 816k/7.98M [00:00<00:02, 3.13MB/
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 4%| | 356k/9.54M [00:00<00:06, 1.44MB/
sub-MGH1_rec-uncombined02_T2starw.nii.gz: 13%|▏| 83.6k/666k [00:00<00:00, 650kB
sub-MGH1_UNIT1.nii.gz: 7%|█ | 1.19M/17.0M [00:00<00:03, 4.21MB/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 26%|▎| 2.09M/7.98M [00:00<00:00, 7.08MB
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 11%| | 1.09M/9.54M [00:00<00:02, 3.85MB
sub-MGH1_rec-uncombined02_T2starw.nii.gz: 53%|▌| 356k/666k [00:00<00:00, 1.50MB
sub-MGH1_UNIT1.nii.gz: 12%|█▊ | 2.10M/17.0M [00:00<00:02, 6.04MB/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 40%|▍| 3.19M/7.98M [00:00<00:00, 8.63MB
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 22%|▏| 2.11M/9.54M [00:00<00:01, 6.20MB
sub-MGH1_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/795k [00:00<?, ?B/s]
sub-MGH1_UNIT1.nii.gz: 20%|██▉ | 3.36M/17.0M [00:00<00:01, 8.44MB/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 56%|▌| 4.49M/7.98M [00:00<00:00, 10.3MB
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 33%|▎| 3.13M/9.54M [00:00<00:00, 7.67MB
sub-MGH1_rec-uncombined01_T2starw.nii.gz: 11%| | 85.2k/795k [00:00<00:01, 626kB
sub-MGH1_UNIT1.nii.gz: 32%|████▊ | 5.41M/17.0M [00:00<00:00, 12.6MB/s]
sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz: 82%|▊| 6.57M/7.98M [00:00<00:00, 14.0MB
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 4.99M/9.54M [00:00<00:00, 11.5MB
sub-MGH1_UNIT1.nii.gz: 41%|██████ | 6.94M/17.0M [00:00<00:00, 13.4MB/s]
sub-MGH1_rec-uncombined01_T2starw.nii.gz: 47%|▍| 374k/795k [00:00<00:00, 1.50MB
sub-MGH1_rec-uncombined01_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 65%|▋| 6.21M/9.54M [00:00<00:00, 11.5MB
sub-MGH1_UNIT1.nii.gz: 49%|███████▎ | 8.27M/17.0M [00:00<00:00, 13.2MB/s]
sub-MGH1_inv-2_part-mag_MP2RAGE.nii.gz: 85%|▊| 8.11M/9.54M [00:00<00:00, 14.0MB
sub-MGH1_UNIT1.nii.gz: 63%|█████████▍ | 10.7M/17.0M [00:01<00:00, 16.2MB/s]
sub-MGH1_rec-uncombined03_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined04_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_UNIT1.nii.gz: 77%|███████████▌ | 13.2M/17.0M [00:01<00:00, 19.1MB/s]
sub-MGH1_rec-uncombined05_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_UNIT1.nii.gz: 98%|██████████████▋| 16.7M/17.0M [00:01<00:00, 24.4MB/s]
sub-MGH1_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/688k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/779k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/771k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined03_T2starw.nii.gz: 14%|▏| 94.0k/688k [00:00<00:00, 689kB
sub-MGH1_rec-uncombined04_T2starw.nii.gz: 11%| | 85.2k/779k [00:00<00:01, 655kB
sub-MGH1_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/789k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/804k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined06_T2starw.nii.gz: 11%| | 85.2k/771k [00:00<00:01, 665kB
sub-MGH1_rec-uncombined04_T2starw.nii.gz: 46%|▍| 357k/779k [00:00<00:00, 1.51MB
sub-MGH1_rec-uncombined03_T2starw.nii.gz: 58%|▌| 400k/688k [00:00<00:00, 1.60MB
sub-MGH1_rec-uncombined05_T2starw.nii.gz: 11%| | 85.2k/789k [00:00<00:01, 569kB
sub-MGH1_rec-uncombined07_T2starw.nii.gz: 10%| | 83.6k/804k [00:00<00:01, 608kB
sub-MGH1_rec-uncombined06_T2starw.nii.gz: 44%|▍| 341k/771k [00:00<00:00, 1.68MB
sub-MGH1_rec-uncombined05_T2starw.nii.gz: 45%|▍| 357k/789k [00:00<00:00, 1.56MB
sub-MGH1_rec-uncombined06_T2starw.nii.gz: 97%|▉| 749k/771k [00:00<00:00, 2.74MB
sub-MGH1_rec-uncombined07_T2starw.nii.gz: 44%|▍| 356k/804k [00:00<00:00, 1.42MB
sub-MGH1_rec-uncombined07_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined06_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined08_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined09_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/957k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/871k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined10_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined08_T2starw.nii.gz: 9%| | 83.6k/957k [00:00<00:01, 649kB
sub-MGH1_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/833k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined09_T2starw.nii.gz: 10%| | 83.6k/871k [00:00<00:01, 656kB
sub-MGH1_rec-uncombined11_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined08_T2starw.nii.gz: 37%|▎| 356k/957k [00:00<00:00, 1.50MB
sub-MGH1_rec-uncombined10_T2starw.nii.gz: 10%| | 83.5k/833k [00:00<00:01, 640kB
sub-MGH1_rec-uncombined09_T2starw.nii.gz: 39%|▍| 339k/871k [00:00<00:00, 1.44MB
sub-MGH1_rec-uncombined10_T2starw.nii.gz: 37%|▎| 305k/833k [00:00<00:00, 1.48MB
sub-MGH1_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/895k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined10_T2starw.nii.gz: 96%|▉| 798k/833k [00:00<00:00, 2.99MB
sub-MGH1_rec-uncombined11_T2starw.nii.gz: 9%| | 83.6k/895k [00:00<00:01, 615kB
sub-MGH1_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/955k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined12_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined11_T2starw.nii.gz: 40%|▍| 356k/895k [00:00<00:00, 1.41MB
sub-MGH1_rec-uncombined13_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined12_T2starw.nii.gz: 9%| | 85.2k/955k [00:00<00:01, 598kB
sub-MGH1_rec-uncombined14_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined12_T2starw.nii.gz: 37%|▎| 357k/955k [00:00<00:00, 1.56MB
sub-MGH1_rec-uncombined12_T2starw.nii.gz: 94%|▉| 901k/955k [00:00<00:00, 3.15MB
sub-MGH1_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/789k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined15_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/783k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined13_T2starw.nii.gz: 11%| | 83.6k/789k [00:00<00:01, 644kB
sub-MGH1_rec-uncombined14_T2starw.nii.gz: 11%| | 83.6k/783k [00:00<00:00, 758kB
sub-MGH1_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/809k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined13_T2starw.nii.gz: 45%|▍| 356k/789k [00:00<00:00, 1.50MB
sub-MGH1_rec-uncombined14_T2starw.nii.gz: 45%|▍| 356k/783k [00:00<00:00, 1.61MB
sub-MGH1_rec-uncombined17_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined15_T2starw.nii.gz: 11%| | 85.2k/809k [00:00<00:01, 583kB
sub-MGH1_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/985k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined15_T2starw.nii.gz: 42%|▍| 340k/809k [00:00<00:00, 1.38MB
sub-MGH1_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/985k [00:00<00:01, 613kB
sub-MGH1_rec-uncombined16_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined18_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/960k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined17_T2starw.nii.gz: 34%|▎| 339k/985k [00:00<00:00, 1.35MB
sub-MGH1_rec-uncombined16_T2starw.nii.gz: 9%| | 83.6k/960k [00:00<00:01, 706kB
sub-MGH1_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/890k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined19_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined16_T2starw.nii.gz: 35%|▎| 339k/960k [00:00<00:00, 1.43MB
sub-MGH1_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/827k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined18_T2starw.nii.gz: 9%| | 83.6k/890k [00:00<00:01, 651kB
sub-MGH1_rec-uncombined20_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined19_T2starw.nii.gz: 10%| | 83.6k/827k [00:00<00:01, 616kB
sub-MGH1_rec-uncombined18_T2starw.nii.gz: 38%|▍| 340k/890k [00:00<00:00, 1.43MB
sub-MGH1_rec-uncombined19_T2starw.nii.gz: 41%|▍| 339k/827k [00:00<00:00, 1.58MB
sub-MGH1_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/922k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MGH1_rec-uncombined19_T2starw.nii.gz: 88%|▉| 730k/827k [00:00<00:00, 2.54MB
sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.44M [00:00<?, ?B/s]
sub-MGH1_rec-uncombined20_T2starw.nii.gz: 9%| | 83.6k/922k [00:00<00:01, 611kB
sub-MGH1_acq-anat_TB1TFL.json: 0%| | 0.00/2.62k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 77.0k/4.44M [00:00<00:09, 481kB/
sub-MGH1_rec-uncombined20_T2starw.nii.gz: 39%|▍| 356k/922k [00:00<00:00, 1.34MB
sub-MGH1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-MGH1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/927k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 349k/4.44M [00:00<00:03, 1.37MB/
sub-MGH1_acq-anat_TB1TFL.nii.gz: 7%|▌ | 67.5k/927k [00:00<00:01, 555kB/s]
sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz: 32%|▎| 1.40M/4.44M [00:00<00:00, 4.41MB
sub-MGH1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.70M [00:00<?, ?B/s]
sub-MGH1_acq-anat_TB1TFL.nii.gz: 37%|██▌ | 339k/927k [00:00<00:00, 1.43MB/s]
sub-MGH1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.1M [00:00<?, ?B/s]
sub-MGH1_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.70M [00:00<00:05, 74
sub-MGH1_acq-coilQaSagSmall_GFactor.nii.gz: 9%| | 356k/3.70M [00:00<00:02, 1.5
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/16.1M [00:00<00:27, 607kB/s
sub-MGH1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH1_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/90.8k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaSagSmall_GFactor.nii.gz: 39%|▍| 1.44M/3.70M [00:00<00:00, 4.
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/16.1M [00:00<00:11, 1.42MB/s
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 6%| | 968k/16.1M [00:00<00:05, 3.14MB/s
sub-MGH1_acq-famp-0.66_TB1DREAM.nii.gz: 94%|▉| 85.2k/90.8k [00:00<00:00, 624kB/
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 18%|▏| 2.93M/16.1M [00:00<00:01, 9.11MB/
sub-MGH1_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.6k [00:00<?, ?B/s]
sub-MGH1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/91.4k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 44%|▍| 7.07M/16.1M [00:00<00:00, 20.3MB/
sub-MGH1_acq-famp-1.5_TB1DREAM.nii.gz: 91%|▉| 83.6k/91.6k [00:00<00:00, 613kB/s
sub-MGH1_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 58%|▌| 9.32M/16.1M [00:00<00:00, 20.8MB/
sub-MGH1_acq-famp_TB1DREAM.nii.gz: 91%|███▋| 83.6k/91.4k [00:00<00:00, 651kB/s]
sub-MGH1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MGH1_acq-coilQaTra_GFactor.nii.gz: 74%|▋| 12.0M/16.1M [00:00<00:00, 22.9MB/
sub-MGH1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MGH1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.8k [00:00<?, ?B/s]
sub-MGH1_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MGH1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/950k [00:00<?, ?B/s]
sub-MGH1_acq-refv-0.66_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.8k [00:00<00:00, 706kB/
sub-MGH1_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/950k [00:00<00:01, 627kB/s]
sub-MGH1_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/97.5k [00:00<?, ?B/s]
sub-MGH1_acq-famp_TB1TFL.nii.gz: 37%|██▌ | 356k/950k [00:00<00:00, 1.55MB/s]
sub-MGH1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH1_acq-refv-1.5_TB1DREAM.nii.gz: 87%|▊| 85.2k/97.5k [00:00<00:00, 669kB/s
sub-MGH2_T2starw.nii.gz: 0%| | 0.00/1.16M [00:00<?, ?B/s]
sub-MGH2_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MGH2_T2starw.nii.gz: 7%|▉ | 83.6k/1.16M [00:00<00:01, 694kB/s]
sub-MGH2_T2starw.nii.gz: 30%|████▏ | 356k/1.16M [00:00<00:00, 1.49MB/s]
sub-MGH1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/96.5k [00:00<?, ?B/s]
sub-MGH2_inv-1_MP2RAGE.json: 0%| | 0.00/1.99k [00:00<?, ?B/s]
sub-MGH2_UNIT1.json: 0%| | 0.00/1.98k [00:00<?, ?B/s]
sub-MGH2_T2starw.nii.gz: 74%|██████████▎ | 884k/1.16M [00:00<00:00, 3.00MB/s]
sub-MGH2_UNIT1.nii.gz: 0%| | 0.00/17.1M [00:00<?, ?B/s]
sub-MGH1_acq-refv_TB1DREAM.nii.gz: 88%|███▌| 85.2k/96.5k [00:00<00:00, 619kB/s]
sub-MGH2_UNIT1.nii.gz: 0%| | 77.0k/17.1M [00:00<00:28, 629kB/s]
sub-MGH2_inv-2_MP2RAGE.json: 0%| | 0.00/1.99k [00:00<?, ?B/s]
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/7.82M [00:00<?, ?B/s]
sub-MGH2_UNIT1.nii.gz: 2%|▎ | 366k/17.1M [00:00<00:11, 1.59MB/s]
sub-MGH2_rec-uncombined01_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/7.82M [00:00<00:11, 687kB/
sub-MGH2_UNIT1.nii.gz: 9%|█▎ | 1.47M/17.1M [00:00<00:03, 5.00MB/s]
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/9.49M [00:00<?, ?B/s]
sub-MGH2_UNIT1.nii.gz: 25%|███▋ | 4.27M/17.1M [00:00<00:01, 13.2MB/s]
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 4%| | 356k/7.82M [00:00<00:05, 1.55MB/
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/9.49M [00:00<00:15, 651kB/
sub-MGH2_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/761k [00:00<?, ?B/s]
sub-MGH2_UNIT1.nii.gz: 49%|███████▍ | 8.44M/17.1M [00:00<00:00, 23.1MB/s]
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 1.28M/7.82M [00:00<00:01, 4.78MB
sub-MGH2_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/591k [00:00<?, ?B/s]
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 4%| | 356k/9.49M [00:00<00:06, 1.51MB/
sub-MGH2_UNIT1.nii.gz: 66%|█████████▉ | 11.3M/17.1M [00:00<00:00, 25.2MB/s]
sub-MGH2_rec-uncombined01_T2starw.nii.gz: 11%| | 83.6k/761k [00:00<00:01, 644kB
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 34%|▎| 2.65M/7.82M [00:00<00:00, 8.23MB
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 12%| | 1.09M/9.49M [00:00<00:02, 3.96MB
sub-MGH2_rec-uncombined02_T2starw.nii.gz: 14%|▏| 85.2k/591k [00:00<00:00, 624kB
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 57%|▌| 4.43M/7.82M [00:00<00:00, 11.8MB
sub-MGH2_UNIT1.nii.gz: 81%|████████████▏ | 13.9M/17.1M [00:00<00:00, 22.6MB/s]
sub-MGH2_rec-uncombined01_T2starw.nii.gz: 47%|▍| 356k/761k [00:00<00:00, 1.48MB
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 22%|▏| 2.11M/9.49M [00:00<00:01, 6.33MB
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 77%|▊| 6.05M/7.82M [00:00<00:00, 13.5MB
sub-MGH2_rec-uncombined02_T2starw.nii.gz: 60%|▌| 357k/591k [00:00<00:00, 1.34MB
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 34%|▎| 3.22M/9.49M [00:00<00:00, 7.69MB
sub-MGH2_UNIT1.nii.gz: 94%|██████████████▏| 16.2M/17.1M [00:01<00:00, 18.3MB/s]
sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz: 95%|▉| 7.40M/7.82M [00:00<00:00, 12.6MB
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 49%|▍| 4.67M/9.49M [00:00<00:00, 10.0MB
sub-MGH2_inv-2_part-mag_MP2RAGE.nii.gz: 91%|▉| 8.62M/9.49M [00:00<00:00, 19.8MB
sub-MGH2_rec-uncombined02_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined03_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined04_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/628k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined05_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined03_T2starw.nii.gz: 14%|▏| 85.2k/628k [00:00<00:00, 616kB
sub-MGH2_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/701k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined06_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/727k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined03_T2starw.nii.gz: 57%|▌| 357k/628k [00:00<00:00, 1.40MB
sub-MGH2_rec-uncombined04_T2starw.nii.gz: 12%| | 83.6k/701k [00:00<00:01, 567kB
sub-MGH2_rec-uncombined05_T2starw.nii.gz: 11%| | 83.5k/727k [00:00<00:00, 699kB
sub-MGH2_rec-uncombined04_T2starw.nii.gz: 51%|▌| 356k/701k [00:00<00:00, 1.43MB
sub-MGH2_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/697k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined05_T2starw.nii.gz: 56%|▌| 407k/727k [00:00<00:00, 1.73MB
sub-MGH2_rec-uncombined07_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined06_T2starw.nii.gz: 12%| | 85.2k/697k [00:00<00:00, 666kB
sub-MGH2_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/714k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined08_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined06_T2starw.nii.gz: 59%|▌| 408k/697k [00:00<00:00, 1.61MB
sub-MGH2_rec-uncombined07_T2starw.nii.gz: 12%| | 85.2k/714k [00:00<00:01, 571kB
sub-MGH2_rec-uncombined09_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined07_T2starw.nii.gz: 48%|▍| 340k/714k [00:00<00:00, 1.31MB
sub-MGH2_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/880k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined08_T2starw.nii.gz: 9%| | 83.6k/880k [00:00<00:01, 607kB
sub-MGH2_rec-uncombined10_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/807k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined08_T2starw.nii.gz: 40%|▍| 356k/880k [00:00<00:00, 1.41MB
sub-MGH2_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/777k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined11_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined09_T2starw.nii.gz: 11%| | 85.2k/807k [00:00<00:01, 580kB
sub-MGH2_rec-uncombined12_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined10_T2starw.nii.gz: 11%| | 83.6k/777k [00:00<00:01, 606kB
sub-MGH2_rec-uncombined09_T2starw.nii.gz: 38%|▍| 306k/807k [00:00<00:00, 1.20MB
sub-MGH2_rec-uncombined10_T2starw.nii.gz: 46%|▍| 356k/777k [00:00<00:00, 1.41MB
sub-MGH2_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/817k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined13_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/861k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined11_T2starw.nii.gz: 10%| | 83.6k/817k [00:00<00:01, 651kB
sub-MGH2_rec-uncombined12_T2starw.nii.gz: 10%| | 83.6k/861k [00:00<00:01, 614kB
sub-MGH2_rec-uncombined11_T2starw.nii.gz: 44%|▍| 356k/817k [00:00<00:00, 1.51MB
sub-MGH2_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/740k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined14_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined12_T2starw.nii.gz: 31%|▎| 271k/861k [00:00<00:00, 1.21MB
sub-MGH2_rec-uncombined13_T2starw.nii.gz: 11%| | 83.6k/740k [00:00<00:01, 611kB
sub-MGH2_rec-uncombined12_T2starw.nii.gz: 87%|▊| 747k/861k [00:00<00:00, 2.70MB
sub-MGH2_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/733k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined13_T2starw.nii.gz: 50%|▌| 373k/740k [00:00<00:00, 1.69MB
sub-MGH2_rec-uncombined15_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined14_T2starw.nii.gz: 11%| | 83.6k/733k [00:00<00:01, 614kB
sub-MGH2_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/734k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined16_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined14_T2starw.nii.gz: 49%|▍| 356k/733k [00:00<00:00, 1.45MB
sub-MGH2_rec-uncombined15_T2starw.nii.gz: 11%| | 83.6k/734k [00:00<00:01, 616kB
sub-MGH2_rec-uncombined17_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/927k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined15_T2starw.nii.gz: 48%|▍| 356k/734k [00:00<00:00, 1.37MB
sub-MGH2_rec-uncombined16_T2starw.nii.gz: 9%| | 83.6k/927k [00:00<00:01, 609kB
sub-MGH2_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/985k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined18_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined16_T2starw.nii.gz: 38%|▍| 356k/927k [00:00<00:00, 1.41MB
sub-MGH2_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/985k [00:00<00:01, 606kB
sub-MGH2_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/856k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined17_T2starw.nii.gz: 34%|▎| 339k/985k [00:00<00:00, 1.34MB
sub-MGH2_rec-uncombined18_T2starw.nii.gz: 10%| | 83.6k/856k [00:00<00:01, 613kB
sub-MGH2_rec-uncombined18_T2starw.nii.gz: 30%|▎| 254k/856k [00:00<00:00, 1.16MB
sub-MGH2_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/828k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined19_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined18_T2starw.nii.gz: 83%|▊| 713k/856k [00:00<00:00, 2.57MB
sub-MGH2_rec-uncombined19_T2starw.nii.gz: 10%| | 83.6k/828k [00:00<00:01, 609kB
sub-MGH2_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/941k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined20_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined19_T2starw.nii.gz: 45%|▍| 373k/828k [00:00<00:00, 1.60MB
sub-MGH2_rec-uncombined20_T2starw.nii.gz: 9%| | 83.6k/941k [00:00<00:01, 650kB
sub-MGH2_acq-anat_TB1TFL.json: 0%| | 0.00/2.62k [00:00<?, ?B/s]
sub-MGH2_rec-uncombined20_T2starw.nii.gz: 38%|▍| 356k/941k [00:00<00:00, 1.51MB
sub-MGH2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/869k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-MGH2_acq-anat_TB1TFL.nii.gz: 10%|▋ | 85.2k/869k [00:00<00:01, 627kB/s]
sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.12M [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-MGH2_acq-anat_TB1TFL.nii.gz: 41%|██▉ | 357k/869k [00:00<00:00, 1.41MB/s]
sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.5k/4.12M [00:00<00:07, 575kB/
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.9M [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.65M [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz: 10%| | 407k/4.12M [00:00<00:02, 1.66MB/
sub-MGH2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/15.9M [00:00<00:25, 649kB/s
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 77.0k/3.65M [00:00<00:06, 56
sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz: 40%|▍| 1.64M/4.12M [00:00<00:00, 5.18MB
sub-MGH2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 6%| | 213k/3.65M [00:00<00:03, 982
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/15.9M [00:00<00:09, 1.74MB/s
sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz: 98%|▉| 4.03M/4.12M [00:00<00:00, 11.5MB
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 20%|▏| 740k/3.65M [00:00<00:01, 2.7
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 10%| | 1.64M/15.9M [00:00<00:02, 5.45MB/
sub-MGH2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/90.8k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 65%|▋| 2.38M/3.65M [00:00<00:00, 8.
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 29%|▎| 4.62M/15.9M [00:00<00:00, 14.0MB/
sub-MGH2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH2_acq-famp-0.66_TB1DREAM.nii.gz: 92%|▉| 83.5k/90.8k [00:00<00:00, 612kB/
sub-MGH2_acq-coilQaSagSmall_GFactor.nii.gz: 100%|█| 3.65M/3.65M [00:00<00:00, 9.
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 38%|▍| 6.12M/15.9M [00:00<00:00, 13.8MB/
sub-MGH2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.9k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 70%|▋| 11.1M/15.9M [00:00<00:00, 25.3MB/
sub-MGH2_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 85.2k/91.9k [00:00<00:00, 660kB/s
sub-MGH2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MGH2_acq-coilQaTra_GFactor.nii.gz: 87%|▊| 13.8M/15.9M [00:00<00:00, 26.1MB/
sub-MGH2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/927k [00:00<?, ?B/s]
sub-MGH2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/91.1k [00:00<?, ?B/s]
sub-MGH2_acq-famp_TB1DREAM.nii.gz: 92%|███▋| 83.6k/91.1k [00:00<00:00, 646kB/s]
sub-MGH2_acq-famp_TB1TFL.nii.gz: 9%|▋ | 85.2k/927k [00:00<00:01, 546kB/s]
sub-MGH2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH2_acq-famp_TB1TFL.json: 0%| | 0.00/2.65k [00:00<?, ?B/s]
sub-MGH2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MGH2_acq-famp_TB1TFL.nii.gz: 39%|██▋ | 357k/927k [00:00<00:00, 1.46MB/s]
sub-MGH2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.6k [00:00<?, ?B/s]
sub-MGH2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/96.1k [00:00<?, ?B/s]
sub-MGH2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/93.5k [00:00<?, ?B/s]
sub-MGH3_T2starw.json: 0%| | 0.00/2.43k [00:00<?, ?B/s]
sub-MGH2_acq-refv-0.66_TB1DREAM.nii.gz: 94%|▉| 83.6k/88.6k [00:00<00:00, 615kB/
sub-MGH2_acq-refv-1.5_TB1DREAM.nii.gz: 80%|▊| 77.0k/96.1k [00:00<00:00, 551kB/s
sub-MGH2_acq-refv_TB1DREAM.nii.gz: 89%|███▌| 83.6k/93.5k [00:00<00:00, 655kB/s]
sub-MGH3_T2starw.nii.gz: 0%| | 0.00/1.33M [00:00<?, ?B/s]
sub-MGH3_inv-1_MP2RAGE.json: 0%| | 0.00/1.98k [00:00<?, ?B/s]
sub-MGH3_T2starw.nii.gz: 6%|▉ | 85.2k/1.33M [00:00<00:01, 654kB/s]
sub-MGH3_UNIT1.json: 0%| | 0.00/1.98k [00:00<?, ?B/s]
sub-MGH3_UNIT1.nii.gz: 0%| | 0.00/17.1M [00:00<?, ?B/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/10.4M [00:00<?, ?B/s]
sub-MGH3_T2starw.nii.gz: 28%|███▊ | 375k/1.33M [00:00<00:00, 1.59MB/s]
sub-MGH3_UNIT1.nii.gz: 0%| | 85.2k/17.1M [00:00<00:22, 806kB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 1%| | 85.2k/10.4M [00:00<00:17, 625kB/
sub-MGH3_rec-uncombined01_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_UNIT1.nii.gz: 2%|▎ | 357k/17.1M [00:00<00:11, 1.58MB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 3%| | 340k/10.4M [00:00<00:07, 1.35MB/
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/8.41M [00:00<?, ?B/s]
sub-MGH3_UNIT1.nii.gz: 8%|█▎ | 1.45M/17.1M [00:00<00:03, 4.77MB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 14%|▏| 1.41M/10.4M [00:00<00:02, 4.42MB
sub-MGH3_inv-2_MP2RAGE.json: 0%| | 0.00/1.98k [00:00<?, ?B/s]
sub-MGH3_UNIT1.nii.gz: 18%|██▋ | 3.06M/17.1M [00:00<00:01, 8.75MB/s]
sub-MGH3_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/795k [00:00<?, ?B/s]
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/8.41M [00:00<00:14, 591kB/
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 33%|▎| 3.44M/10.4M [00:00<00:00, 9.75MB
sub-MGH3_UNIT1.nii.gz: 36%|█████▍ | 6.21M/17.1M [00:00<00:00, 16.5MB/s]
sub-MGH3_rec-uncombined01_T2starw.nii.gz: 11%| | 83.6k/795k [00:00<00:01, 621kB
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 47%|▍| 4.94M/10.4M [00:00<00:00, 11.6MB
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 5%| | 390k/8.41M [00:00<00:05, 1.51MB/
sub-MGH3_UNIT1.nii.gz: 50%|███████▌ | 8.59M/17.1M [00:00<00:00, 19.1MB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 67%|▋| 7.01M/10.4M [00:00<00:00, 14.7MB
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 13%|▏| 1.10M/8.41M [00:00<00:02, 3.70MB
sub-MGH3_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/632k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined01_T2starw.nii.gz: 45%|▍| 356k/795k [00:00<00:00, 1.43MB
sub-MGH3_UNIT1.nii.gz: 62%|█████████▏ | 10.5M/17.1M [00:00<00:00, 18.9MB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 83%|▊| 8.64M/10.4M [00:00<00:00, 15.4MB
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 24%|▏| 2.05M/8.41M [00:00<00:01, 5.48MB
sub-MGH3_rec-uncombined02_T2starw.nii.gz: 13%|▏| 85.2k/632k [00:00<00:00, 643kB
sub-MGH3_UNIT1.nii.gz: 73%|██████████▉ | 12.4M/17.1M [00:00<00:00, 17.5MB/s]
sub-MGH3_inv-2_part-mag_MP2RAGE.nii.gz: 98%|▉| 10.2M/10.4M [00:00<00:00, 15.3MB
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 44%|▍| 3.71M/8.41M [00:00<00:00, 8.84MB
sub-MGH3_rec-uncombined02_T2starw.nii.gz: 57%|▌| 357k/632k [00:00<00:00, 1.50MB
sub-MGH3_UNIT1.nii.gz: 83%|████████████▍ | 14.2M/17.1M [00:01<00:00, 17.4MB/s]
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 63%|▋| 5.30M/8.41M [00:00<00:00, 11.2MB
sub-MGH3_rec-uncombined02_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_UNIT1.nii.gz: 93%|█████████████▉ | 15.9M/17.1M [00:01<00:00, 16.7MB/s]
sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz: 87%|▊| 7.28M/8.41M [00:00<00:00, 14.1MB
sub-MGH3_rec-uncombined03_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/683k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined04_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined06_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined03_T2starw.nii.gz: 12%| | 83.6k/683k [00:00<00:00, 649kB
sub-MGH3_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/758k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/768k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined03_T2starw.nii.gz: 52%|▌| 356k/683k [00:00<00:00, 1.50MB
sub-MGH3_rec-uncombined04_T2starw.nii.gz: 11%| | 83.6k/758k [00:00<00:01, 613kB
sub-MGH3_rec-uncombined05_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined05_T2starw.nii.gz: 11%| | 83.6k/768k [00:00<00:01, 586kB
sub-MGH3_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/751k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined04_T2starw.nii.gz: 47%|▍| 356k/758k [00:00<00:00, 1.42MB
sub-MGH3_rec-uncombined05_T2starw.nii.gz: 46%|▍| 356k/768k [00:00<00:00, 1.44MB
sub-MGH3_rec-uncombined06_T2starw.nii.gz: 11%| | 85.2k/751k [00:00<00:01, 567kB
sub-MGH3_rec-uncombined07_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined05_T2starw.nii.gz: 100%|█| 768k/768k [00:00<00:00, 2.50MB
sub-MGH3_rec-uncombined06_T2starw.nii.gz: 48%|▍| 357k/751k [00:00<00:00, 1.56MB
sub-MGH3_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/988k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined08_T2starw.nii.gz: 8%| | 83.6k/988k [00:00<00:01, 656kB
sub-MGH3_rec-uncombined08_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined09_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined08_T2starw.nii.gz: 36%|▎| 356k/988k [00:00<00:00, 1.51MB
sub-MGH3_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/808k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/860k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined07_T2starw.nii.gz: 10%| | 83.6k/808k [00:00<00:01, 700kB
sub-MGH3_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/824k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined10_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined07_T2starw.nii.gz: 44%|▍| 356k/808k [00:00<00:00, 1.48MB
sub-MGH3_rec-uncombined09_T2starw.nii.gz: 10%| | 83.6k/860k [00:00<00:01, 574kB
sub-MGH3_rec-uncombined10_T2starw.nii.gz: 10%| | 83.6k/824k [00:00<00:01, 652kB
sub-MGH3_rec-uncombined11_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined09_T2starw.nii.gz: 32%|▎| 272k/860k [00:00<00:00, 1.16MB
sub-MGH3_rec-uncombined10_T2starw.nii.gz: 43%|▍| 356k/824k [00:00<00:00, 1.50MB
sub-MGH3_rec-uncombined12_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/910k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/995k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined11_T2starw.nii.gz: 8%| | 77.0k/910k [00:00<00:01, 631kB
sub-MGH3_rec-uncombined13_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined12_T2starw.nii.gz: 8%| | 83.6k/995k [00:00<00:01, 610kB
sub-MGH3_rec-uncombined14_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined11_T2starw.nii.gz: 38%|▍| 349k/910k [00:00<00:00, 1.45MB
sub-MGH3_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/776k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined12_T2starw.nii.gz: 36%|▎| 356k/995k [00:00<00:00, 1.42MB
sub-MGH3_rec-uncombined13_T2starw.nii.gz: 11%| | 85.2k/776k [00:00<00:01, 624kB
sub-MGH3_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/799k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined13_T2starw.nii.gz: 46%|▍| 357k/776k [00:00<00:00, 1.43MB
sub-MGH3_rec-uncombined15_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/820k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined14_T2starw.nii.gz: 10%| | 83.6k/799k [00:00<00:01, 613kB
sub-MGH3_rec-uncombined16_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined15_T2starw.nii.gz: 10%| | 78.0k/820k [00:00<00:01, 633kB
sub-MGH3_rec-uncombined14_T2starw.nii.gz: 25%|▎| 204k/799k [00:00<00:00, 923kB/
sub-MGH3_rec-uncombined14_T2starw.nii.gz: 91%|▉| 730k/799k [00:00<00:00, 2.68MB
sub-MGH3_rec-uncombined15_T2starw.nii.gz: 45%|▍| 366k/820k [00:00<00:00, 1.54MB
sub-MGH3_rec-uncombined17_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-MGH3_rec-uncombined18_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined16_T2starw.nii.gz: 8%| | 85.2k/1.07M [00:00<00:01, 621k
sub-MGH3_rec-uncombined16_T2starw.nii.gz: 34%|▎| 374k/1.07M [00:00<00:00, 1.49M
sub-MGH3_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-MGH3_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/994k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MGH3_rec-uncombined19_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined17_T2starw.nii.gz: 8%| | 83.5k/1.08M [00:00<00:01, 642k
sub-MGH3_rec-uncombined19_T2starw.nii.gz: 8%| | 83.5k/994k [00:00<00:01, 614kB
sub-MGH3_rec-uncombined18_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 607k
sub-MGH3_rec-uncombined17_T2starw.nii.gz: 37%|▎| 407k/1.08M [00:00<00:00, 1.72M
sub-MGH3_rec-uncombined19_T2starw.nii.gz: 36%|▎| 356k/994k [00:00<00:00, 1.42MB
sub-MGH3_rec-uncombined18_T2starw.nii.gz: 37%|▎| 373k/0.99M [00:00<00:00, 1.48M
sub-MGH3_rec-uncombined20_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-MGH3_rec-uncombined19_T2starw.nii.gz: 94%|▉| 935k/994k [00:00<00:00, 3.11MB
sub-MGH3_rec-uncombined18_T2starw.nii.gz: 94%|▉| 952k/0.99M [00:00<00:00, 3.00M
sub-MGH3_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 659k
sub-MGH3_acq-anat_TB1TFL.json: 0%| | 0.00/2.61k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-MGH3_rec-uncombined20_T2starw.nii.gz: 36%|▎| 390k/1.05M [00:00<00:00, 1.55M
sub-MGH3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/966k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.52M [00:00<?, ?B/s]
sub-MGH3_acq-anat_TB1TFL.nii.gz: 9%|▌ | 83.5k/966k [00:00<00:01, 617kB/s]
sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.52M [00:00<00:07, 653kB/
sub-MGH3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-MGH3_acq-anat_TB1TFL.nii.gz: 21%|█▋ | 203k/966k [00:00<00:00, 896kB/s]
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.65M [00:00<?, ?B/s]
sub-MGH3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.52M [00:00<00:03, 1.42MB/
sub-MGH3_acq-anat_TB1TFL.nii.gz: 86%|██████ | 832k/966k [00:00<00:00, 3.14MB/s]
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 94.0k/3.65M [00:00<00:05, 70
sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz: 32%|▎| 1.46M/4.52M [00:00<00:00, 4.95MB
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 11%| | 400k/3.65M [00:00<00:02, 1.6
sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz: 96%|▉| 4.34M/4.52M [00:00<00:00, 13.4MB
sub-MGH3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.8M [00:00<?, ?B/s]
sub-MGH3_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 24%|▏| 910k/3.65M [00:00<00:01, 2.4
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 0%| | 83.6k/16.8M [00:00<00:25, 700kB/s
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 39%|▍| 1.44M/3.65M [00:00<00:00, 3.
sub-MGH3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/91.4k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/16.8M [00:00<00:11, 1.50MB/s
sub-MGH3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 56%|▌| 2.05M/3.65M [00:00<00:00, 3.
sub-MGH3_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-MGH3_acq-famp-0.66_TB1DREAM.nii.gz: 93%|▉| 85.2k/91.4k [00:00<00:00, 669kB/
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.28M/16.8M [00:00<00:03, 4.31MB/
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 68%|▋| 2.50M/3.65M [00:00<00:00, 3.
sub-MGH3_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 85.2k/92.1k [00:00<00:00, 664kB/s
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 21%|▏| 3.60M/16.8M [00:00<00:01, 11.1MB/
sub-MGH3_acq-coilQaSagSmall_GFactor.nii.gz: 84%|▊| 3.08M/3.65M [00:00<00:00, 4.
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 42%|▍| 7.05M/16.8M [00:00<00:00, 19.2MB/
sub-MGH3_acq-famp_TB1TFL.json: 0%| | 0.00/2.65k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 55%|▌| 9.30M/16.8M [00:00<00:00, 20.6MB/
sub-MGH3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/91.7k [00:00<?, ?B/s]
sub-MGH3_acq-coilQaTra_GFactor.nii.gz: 82%|▊| 13.9M/16.8M [00:00<00:00, 28.9MB/
sub-MGH3_acq-famp_TB1DREAM.nii.gz: 91%|███▋| 83.6k/91.7k [00:00<00:00, 654kB/s]
sub-MGH3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/970k [00:00<?, ?B/s]
sub-MGH3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MGH3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/94.2k [00:00<?, ?B/s]
sub-MGH3_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/970k [00:00<00:01, 616kB/s]
sub-MGH3_acq-refv-0.66_TB1DREAM.nii.gz: 89%|▉| 83.6k/94.2k [00:00<00:00, 655kB/
sub-MGH3_acq-famp_TB1TFL.nii.gz: 37%|██▌ | 356k/970k [00:00<00:00, 1.62MB/s]
sub-MGH3_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MGH3_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/98.6k [00:00<?, ?B/s]
sub-MGH3_acq-famp_TB1TFL.nii.gz: 80%|█████▋ | 781k/970k [00:00<00:00, 2.73MB/s]
sub-MGH3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MGH3_acq-refv-1.5_TB1DREAM.nii.gz: 78%|▊| 77.0k/98.6k [00:00<00:00, 677kB/s
sub-MGH3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/97.5k [00:00<?, ?B/s]
sub-MNI1_T2starw.json: 0%| | 0.00/2.38k [00:00<?, ?B/s]
sub-MNI1_UNIT1.json: 0%| | 0.00/2.02k [00:00<?, ?B/s]
sub-MNI1_T2starw.nii.gz: 0%| | 0.00/1.33M [00:00<?, ?B/s]
sub-MGH3_acq-refv_TB1DREAM.nii.gz: 86%|███▍| 83.6k/97.5k [00:00<00:00, 551kB/s]
sub-MNI1_UNIT1.nii.gz: 0%| | 0.00/25.9M [00:00<?, ?B/s]
sub-MNI1_T2starw.nii.gz: 6%|▊ | 83.6k/1.33M [00:00<00:02, 648kB/s]
sub-MNI1_inv-1_MP2RAGE.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MNI1_T2starw.nii.gz: 26%|███▋ | 356k/1.33M [00:00<00:00, 1.51MB/s]
sub-MNI1_UNIT1.nii.gz: 0%| | 83.6k/25.9M [00:00<00:44, 609kB/s]
sub-MNI1_inv-2_MP2RAGE.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MNI1_UNIT1.nii.gz: 1%|▏ | 356k/25.9M [00:00<00:18, 1.42MB/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.1M [00:00<?, ?B/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/17.1M [00:00<00:27, 657kB/
sub-MNI1_UNIT1.nii.gz: 6%|▊ | 1.46M/25.9M [00:00<00:05, 4.57MB/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.1M [00:00<?, ?B/s]
sub-MNI1_rec-uncombined01_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_UNIT1.nii.gz: 12%|█▊ | 3.04M/25.9M [00:00<00:02, 8.35MB/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 356k/17.1M [00:00<00:11, 1.52MB/
sub-MNI1_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/822k [00:00<?, ?B/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/19.1M [00:00<00:32, 614kB/
sub-MNI1_UNIT1.nii.gz: 28%|████▏ | 7.35M/25.9M [00:00<00:00, 19.8MB/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.21M/17.1M [00:00<00:03, 4.45MB
sub-MNI1_rec-uncombined01_T2starw.nii.gz: 10%| | 85.2k/822k [00:00<00:01, 614kB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 356k/19.1M [00:00<00:13, 1.42MB/
sub-MNI1_UNIT1.nii.gz: 38%|█████▋ | 9.91M/25.9M [00:00<00:00, 22.0MB/s]
sub-MNI1_rec-uncombined02_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 2.75M/17.1M [00:00<00:01, 8.59MB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 5%| | 1.05M/19.1M [00:00<00:05, 3.63MB
sub-MNI1_rec-uncombined01_T2starw.nii.gz: 43%|▍| 357k/822k [00:00<00:00, 1.42MB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.08M/17.1M [00:00<00:01, 10.4MB
sub-MNI1_UNIT1.nii.gz: 47%|███████ | 12.2M/25.9M [00:00<00:00, 19.2MB/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 12%| | 2.23M/19.1M [00:00<00:02, 6.66MB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 31%|▎| 5.34M/17.1M [00:00<00:01, 11.1MB
sub-MNI1_UNIT1.nii.gz: 55%|████████▏ | 14.2M/25.9M [00:01<00:00, 18.0MB/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 19%|▏| 3.65M/19.1M [00:00<00:01, 9.37MB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 43%|▍| 7.33M/17.1M [00:00<00:00, 14.1MB
sub-MNI1_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/766k [00:00<?, ?B/s]
sub-MNI1_UNIT1.nii.gz: 62%|█████████▎ | 16.1M/25.9M [00:01<00:00, 18.5MB/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 29%|▎| 5.54M/19.1M [00:00<00:01, 12.7MB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 54%|▌| 9.25M/17.1M [00:00<00:00, 16.0MB
sub-MNI1_UNIT1.nii.gz: 70%|██████████▍ | 18.1M/25.9M [00:01<00:00, 19.3MB/s]
sub-MNI1_rec-uncombined03_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 39%|▍| 7.42M/19.1M [00:00<00:00, 13.9MB
sub-MNI1_rec-uncombined02_T2starw.nii.gz: 11%| | 83.6k/766k [00:00<00:01, 582kB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 63%|▋| 10.8M/17.1M [00:00<00:00, 15.7MB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 48%|▍| 9.17M/19.1M [00:00<00:00, 15.2MB
sub-MNI1_UNIT1.nii.gz: 77%|███████████▌ | 20.1M/25.9M [00:01<00:00, 17.9MB/s]
sub-MNI1_rec-uncombined02_T2starw.nii.gz: 49%|▍| 374k/766k [00:00<00:00, 1.65MB
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 74%|▋| 12.7M/17.1M [00:01<00:00, 16.8MB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 56%|▌| 10.7M/19.1M [00:01<00:00, 14.8MB
sub-MNI1_UNIT1.nii.gz: 84%|████████████▌ | 21.8M/25.9M [00:01<00:00, 16.9MB/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 83%|▊| 14.3M/17.1M [00:01<00:00, 16.1MB
sub-MNI1_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/843k [00:00<?, ?B/s]
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 66%|▋| 12.7M/19.1M [00:01<00:00, 16.7MB
sub-MNI1_UNIT1.nii.gz: 92%|█████████████▊ | 23.8M/25.9M [00:01<00:00, 17.9MB/s]
sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz: 95%|▉| 16.3M/17.1M [00:01<00:00, 17.4MB
sub-MNI1_rec-uncombined03_T2starw.nii.gz: 10%| | 83.6k/843k [00:00<00:01, 652kB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 75%|▋| 14.3M/19.1M [00:01<00:00, 16.6MB
sub-MNI1_UNIT1.nii.gz: 99%|██████████████▊| 25.6M/25.9M [00:01<00:00, 17.9MB/s]
sub-MNI1_rec-uncombined04_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined03_T2starw.nii.gz: 44%|▍| 373k/843k [00:00<00:00, 1.86MB
sub-MNI1_inv-2_part-mag_MP2RAGE.nii.gz: 83%|▊| 15.9M/19.1M [00:01<00:00, 16.7MB
sub-MNI1_rec-uncombined03_T2starw.nii.gz: 93%|▉| 781k/843k [00:00<00:00, 2.83MB
sub-MNI1_rec-uncombined05_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/914k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined06_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined04_T2starw.nii.gz: 9%| | 83.6k/914k [00:00<00:01, 590kB
sub-MNI1_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/865k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/828k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined07_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined04_T2starw.nii.gz: 37%|▎| 339k/914k [00:00<00:00, 1.33MB
sub-MNI1_rec-uncombined05_T2starw.nii.gz: 10%| | 83.6k/865k [00:00<00:01, 652kB
sub-MNI1_rec-uncombined06_T2starw.nii.gz: 10%| | 83.6k/828k [00:00<00:01, 655kB
sub-MNI1_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/891k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined05_T2starw.nii.gz: 41%|▍| 356k/865k [00:00<00:00, 1.52MB
sub-MNI1_rec-uncombined06_T2starw.nii.gz: 43%|▍| 356k/828k [00:00<00:00, 1.52MB
sub-MNI1_rec-uncombined08_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined07_T2starw.nii.gz: 9%| | 83.6k/891k [00:00<00:01, 559kB
sub-MNI1_rec-uncombined07_T2starw.nii.gz: 40%|▍| 356k/891k [00:00<00:00, 1.49MB
sub-MNI1_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/979k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined08_T2starw.nii.gz: 9%| | 83.6k/979k [00:00<00:01, 611kB
sub-MNI1_rec-uncombined09_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/860k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/894k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined08_T2starw.nii.gz: 36%|▎| 356k/979k [00:00<00:00, 1.42MB
sub-MNI1_rec-uncombined10_T2starw.nii.gz: 10%| | 83.6k/860k [00:00<00:01, 779kB
sub-MNI1_rec-uncombined09_T2starw.nii.gz: 9%| | 83.6k/894k [00:00<00:01, 668kB
sub-MNI1_rec-uncombined11_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined10_T2starw.nii.gz: 37%|▎| 322k/860k [00:00<00:00, 1.71MB
sub-MNI1_rec-uncombined09_T2starw.nii.gz: 38%|▍| 339k/894k [00:00<00:00, 1.40MB
sub-MNI1_rec-uncombined10_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined10_T2starw.nii.gz: 95%|▉| 816k/860k [00:00<00:00, 3.21MB
sub-MNI1_rec-uncombined12_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/941k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined13_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/810k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined11_T2starw.nii.gz: 9%| | 83.6k/941k [00:00<00:01, 642kB
sub-MNI1_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/800k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/975k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined13_T2starw.nii.gz: 10%| | 83.6k/810k [00:00<00:01, 613kB
sub-MNI1_rec-uncombined11_T2starw.nii.gz: 34%|▎| 322k/941k [00:00<00:00, 1.34MB
sub-MNI1_rec-uncombined14_T2starw.nii.gz: 11%| | 85.2k/800k [00:00<00:01, 663kB
sub-MNI1_rec-uncombined12_T2starw.nii.gz: 10%| | 94.0k/975k [00:00<00:01, 689kB
sub-MNI1_rec-uncombined14_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined13_T2starw.nii.gz: 44%|▍| 356k/810k [00:00<00:00, 1.41MB
sub-MNI1_rec-uncombined14_T2starw.nii.gz: 30%|▎| 238k/800k [00:00<00:00, 1.13MB
sub-MNI1_rec-uncombined12_T2starw.nii.gz: 41%|▍| 400k/975k [00:00<00:00, 1.47MB
sub-MNI1_rec-uncombined14_T2starw.nii.gz: 98%|▉| 783k/800k [00:00<00:00, 3.03MB
sub-MNI1_rec-uncombined15_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined16_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/777k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined17_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/940k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined15_T2starw.nii.gz: 11%| | 83.6k/777k [00:00<00:01, 646kB
sub-MNI1_rec-uncombined18_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined16_T2starw.nii.gz: 8%| | 77.0k/940k [00:00<00:01, 604kB
sub-MNI1_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/894k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined15_T2starw.nii.gz: 46%|▍| 356k/777k [00:00<00:00, 1.50MB
sub-MNI1_rec-uncombined16_T2starw.nii.gz: 39%|▍| 366k/940k [00:00<00:00, 1.55MB
sub-MNI1_rec-uncombined18_T2starw.nii.gz: 9%| | 83.6k/894k [00:00<00:01, 640kB
sub-MNI1_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined19_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined18_T2starw.nii.gz: 38%|▍| 340k/894k [00:00<00:00, 1.36MB
sub-MNI1_rec-uncombined17_T2starw.nii.gz: 9%| | 83.6k/968k [00:00<00:01, 667kB
sub-MNI1_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/813k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined20_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined17_T2starw.nii.gz: 37%|▎| 356k/968k [00:00<00:00, 1.38MB
sub-MNI1_rec-uncombined19_T2starw.nii.gz: 10%| | 83.6k/813k [00:00<00:01, 589kB
sub-MNI1_acq-anat_TB1TFL.json: 0%| | 0.00/2.60k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/924k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined19_T2starw.nii.gz: 42%|▍| 339k/813k [00:00<00:00, 1.46MB
sub-MNI1_rec-uncombined20_T2starw.nii.gz: 9%| | 83.6k/924k [00:00<00:01, 611kB
sub-MNI1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/943k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MNI1_rec-uncombined20_T2starw.nii.gz: 38%|▍| 356k/924k [00:00<00:00, 1.43MB
sub-MNI1_acq-anat_TB1TFL.nii.gz: 9%|▋ | 85.2k/943k [00:00<00:01, 660kB/s]
sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.59M [00:00<?, ?B/s]
sub-MNI1_acq-anat_TB1TFL.nii.gz: 23%|█▋ | 221k/943k [00:00<00:00, 1.04MB/s]
sub-MNI1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.59M [00:00<00:07, 613kB/
sub-MNI1_acq-anat_TB1TFL.nii.gz: 61%|████▎ | 578k/943k [00:00<00:00, 2.07MB/s]
sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.59M [00:00<00:02, 1.52MB/
sub-MNI1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.68M [00:00<?, ?B/s]
sub-MNI1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz: 21%|▏| 968k/4.59M [00:00<00:01, 3.37MB/
sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz: 64%|▋| 2.95M/4.59M [00:00<00:00, 9.67MB
sub-MNI1_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 85.2k/3.68M [00:00<00:06, 62
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.2M [00:00<?, ?B/s]
sub-MNI1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaSagSmall_GFactor.nii.gz: 9%| | 340k/3.68M [00:00<00:02, 1.6
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/16.2M [00:00<00:26, 648kB/s
sub-MNI1_acq-coilQaSagSmall_GFactor.nii.gz: 19%|▏| 731k/3.68M [00:00<00:01, 2.5
sub-MNI1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.9k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/16.2M [00:00<00:11, 1.51MB/s
sub-MNI1_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaSagSmall_GFactor.nii.gz: 51%|▌| 1.88M/3.68M [00:00<00:00, 5.
sub-MNI1_acq-famp-0.66_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.9k [00:00<00:00, 617kB/
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 7%| | 1.08M/16.2M [00:00<00:04, 3.71MB/
sub-MNI1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.3k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 21%|▏| 3.40M/16.2M [00:00<00:01, 10.8MB/
sub-MNI1_acq-famp_TB1DREAM.nii.gz: 92%|███▋| 85.2k/92.3k [00:00<00:00, 806kB/s]
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 46%|▍| 7.37M/16.2M [00:00<00:00, 21.0MB/
sub-MNI1_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-MNI1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 64%|▋| 10.4M/16.2M [00:00<00:00, 23.7MB/
sub-MNI1_acq-famp-1.5_TB1DREAM.nii.gz: 93%|▉| 85.2k/92.1k [00:00<00:00, 668kB/s
sub-MNI1_acq-famp_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-MNI1_acq-coilQaTra_GFactor.nii.gz: 82%|▊| 13.3M/16.2M [00:00<00:00, 24.3MB/
sub-MNI1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/952k [00:00<?, ?B/s]
sub-MNI1_acq-famp_TB1TFL.nii.gz: 7%|▍ | 66.6k/952k [00:00<00:01, 528kB/s]
sub-MNI1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MNI1_acq-famp_TB1TFL.nii.gz: 34%|██▎ | 322k/952k [00:00<00:00, 1.45MB/s]
sub-MNI1_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MNI1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.9k [00:00<?, ?B/s]
sub-MNI1_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/95.6k [00:00<?, ?B/s]
sub-MNI1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI1_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 88.9k/88.9k [00:00<00:00, 666kB/
sub-MNI1_acq-refv-1.5_TB1DREAM.nii.gz: 87%|▊| 83.5k/95.6k [00:00<00:00, 633kB/s
sub-MNI1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/94.5k [00:00<?, ?B/s]
sub-MNI2_T2starw.json: 0%| | 0.00/2.38k [00:00<?, ?B/s]
sub-MNI1_acq-refv_TB1DREAM.nii.gz: 88%|███▌| 83.5k/94.5k [00:00<00:00, 604kB/s]
sub-MNI2_T2starw.nii.gz: 0%| | 0.00/1.18M [00:00<?, ?B/s]
sub-MNI2_inv-1_MP2RAGE.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 0%| | 0.00/26.1M [00:00<?, ?B/s]
sub-MNI2_T2starw.nii.gz: 7%|▉ | 83.6k/1.18M [00:00<00:01, 646kB/s]
sub-MNI2_UNIT1.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 0%| | 85.2k/26.1M [00:00<00:36, 742kB/s]
sub-MNI2_T2starw.nii.gz: 28%|███▉ | 339k/1.18M [00:00<00:00, 1.43MB/s]
sub-MNI2_rec-uncombined01_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/16.7M [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 1%|▏ | 357k/26.1M [00:00<00:19, 1.39MB/s]
sub-MNI2_inv-2_MP2RAGE.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 6%|▊ | 1.45M/26.1M [00:00<00:05, 4.57MB/s]
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/16.7M [00:00<00:26, 650kB/
sub-MNI2_UNIT1.nii.gz: 17%|██▌ | 4.48M/26.1M [00:00<00:01, 12.3MB/s]
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 351k/16.7M [00:00<00:12, 1.40MB/
sub-MNI2_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/718k [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 33%|████▉ | 8.56M/26.1M [00:00<00:00, 21.3MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/14.8M [00:00<?, ?B/s]
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 7%| | 1.16M/16.7M [00:00<00:03, 4.08MB
sub-MNI2_rec-uncombined02_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_UNIT1.nii.gz: 41%|██████▏ | 10.8M/26.1M [00:00<00:00, 21.6MB/s]
sub-MNI2_rec-uncombined01_T2starw.nii.gz: 12%| | 83.5k/718k [00:00<00:01, 610kB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 2.52M/16.7M [00:00<00:01, 7.66MB
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 83.6k/14.8M [00:00<00:25, 613kB/
sub-MNI2_UNIT1.nii.gz: 51%|███████▌ | 13.3M/26.1M [00:00<00:00, 22.7MB/s]
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 30%|▎| 4.99M/16.7M [00:00<00:00, 13.7MB
sub-MNI2_rec-uncombined01_T2starw.nii.gz: 57%|▌| 407k/718k [00:00<00:00, 1.63MB
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 339k/14.8M [00:00<00:12, 1.26MB/
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 39%|▍| 6.57M/16.7M [00:00<00:00, 14.6MB
sub-MNI2_UNIT1.nii.gz: 59%|████████▉ | 15.5M/26.1M [00:00<00:00, 21.2MB/s]
sub-MNI2_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/775k [00:00<?, ?B/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 8%| | 1.23M/14.8M [00:00<00:03, 4.18MB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 8.60M/16.7M [00:00<00:00, 16.7MB
sub-MNI2_UNIT1.nii.gz: 68%|██████████▏ | 17.7M/26.1M [00:01<00:00, 21.4MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 18%|▏| 2.60M/14.8M [00:00<00:01, 7.63MB
sub-MNI2_rec-uncombined03_T2starw.nii.gz: 11%| | 83.6k/775k [00:00<00:01, 651kB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 63%|▋| 10.4M/16.7M [00:00<00:00, 17.4MB
sub-MNI2_UNIT1.nii.gz: 76%|███████████▎ | 19.8M/26.1M [00:01<00:00, 20.7MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 30%|▎| 4.45M/14.8M [00:00<00:00, 11.4MB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 74%|▋| 12.3M/16.7M [00:00<00:00, 18.1MB
sub-MNI2_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/723k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined03_T2starw.nii.gz: 46%|▍| 356k/775k [00:00<00:00, 1.51MB
sub-MNI2_UNIT1.nii.gz: 83%|████████████▌ | 21.8M/26.1M [00:01<00:00, 20.0MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 42%|▍| 6.18M/14.8M [00:00<00:00, 13.6MB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 84%|▊| 14.1M/16.7M [00:01<00:00, 16.7MB
sub-MNI2_rec-uncombined02_T2starw.nii.gz: 11%| | 77.0k/723k [00:00<00:01, 563kB
sub-MNI2_UNIT1.nii.gz: 91%|█████████████▋ | 23.8M/26.1M [00:01<00:00, 18.3MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 51%|▌| 7.56M/14.8M [00:00<00:00, 13.8MB
sub-MNI2_inv-2_part-mag_MP2RAGE.nii.gz: 96%|▉| 15.9M/16.7M [00:01<00:00, 17.5MB
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 61%|▌| 9.10M/14.8M [00:00<00:00, 14.1MB
sub-MNI2_rec-uncombined02_T2starw.nii.gz: 53%|▌| 383k/723k [00:00<00:00, 1.48MB
sub-MNI2_UNIT1.nii.gz: 98%|██████████████▋| 25.5M/26.1M [00:01<00:00, 17.2MB/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 72%|▋| 10.6M/14.8M [00:01<00:00, 14.7MB
sub-MNI2_rec-uncombined03_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz: 99%|▉| 14.7M/14.8M [00:01<00:00, 23.1MB
sub-MNI2_rec-uncombined04_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/782k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined06_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined05_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined04_T2starw.nii.gz: 11%| | 83.6k/782k [00:00<00:01, 652kB
sub-MNI2_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/767k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined04_T2starw.nii.gz: 45%|▍| 356k/782k [00:00<00:00, 1.51MB
sub-MNI2_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/760k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined07_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined05_T2starw.nii.gz: 11%| | 85.2k/767k [00:00<00:01, 626kB
sub-MNI2_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/784k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined06_T2starw.nii.gz: 11%| | 85.2k/760k [00:00<00:01, 661kB
sub-MNI2_rec-uncombined05_T2starw.nii.gz: 49%|▍| 374k/767k [00:00<00:00, 1.80MB
sub-MNI2_rec-uncombined08_T2starw.nii.gz: 11%| | 83.6k/784k [00:00<00:01, 613kB
sub-MNI2_rec-uncombined06_T2starw.nii.gz: 47%|▍| 357k/760k [00:00<00:00, 1.51MB
sub-MNI2_rec-uncombined08_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined08_T2starw.nii.gz: 32%|▎| 254k/784k [00:00<00:00, 1.06MB
sub-MNI2_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/783k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined09_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined07_T2starw.nii.gz: 11%| | 87.4k/783k [00:00<00:01, 656kB
sub-MNI2_rec-uncombined07_T2starw.nii.gz: 52%|▌| 409k/783k [00:00<00:00, 1.75MB
sub-MNI2_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/781k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/782k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined11_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined10_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined09_T2starw.nii.gz: 12%| | 94.0k/781k [00:00<00:00, 714kB
sub-MNI2_rec-uncombined10_T2starw.nii.gz: 11%| | 83.6k/782k [00:00<00:01, 585kB
sub-MNI2_rec-uncombined09_T2starw.nii.gz: 51%|▌| 400k/781k [00:00<00:00, 1.71MB
sub-MNI2_rec-uncombined10_T2starw.nii.gz: 48%|▍| 373k/782k [00:00<00:00, 1.47MB
sub-MNI2_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/845k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined12_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined11_T2starw.nii.gz: 10%| | 83.5k/845k [00:00<00:01, 598kB
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/831k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined11_T2starw.nii.gz: 42%|▍| 356k/845k [00:00<00:00, 1.43MB
sub-MNI2_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/727k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 9%| | 78.6k/831k [00:00<00:01, 540kB
sub-MNI2_rec-uncombined13_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined14_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined13_T2starw.nii.gz: 11%| | 83.6k/727k [00:00<00:01, 652kB
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 22%|▏| 180k/831k [00:00<00:01, 653kB/
sub-MNI2_rec-uncombined13_T2starw.nii.gz: 51%|▌| 373k/727k [00:00<00:00, 1.59MB
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 34%|▎| 282k/831k [00:00<00:00, 694kB/
sub-MNI2_rec-uncombined15_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined16_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/730k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 50%|▌| 418k/831k [00:00<00:00, 809kB/
sub-MNI2_rec-uncombined14_T2starw.nii.gz: 11%| | 83.6k/730k [00:00<00:01, 609kB
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 65%|▋| 537k/831k [00:00<00:00, 831kB/
sub-MNI2_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/667k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined17_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 75%|▋| 622k/831k [00:00<00:00, 842kB/
sub-MNI2_rec-uncombined14_T2starw.nii.gz: 53%|▌| 390k/730k [00:00<00:00, 1.50MB
sub-MNI2_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/780k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined15_T2starw.nii.gz: 13%|▏| 83.6k/667k [00:00<00:00, 616kB
sub-MNI2_rec-uncombined12_T2starw.nii.gz: 93%|▉| 775k/831k [00:00<00:00, 987kB/
sub-MNI2_rec-uncombined16_T2starw.nii.gz: 11%| | 83.6k/780k [00:00<00:01, 613kB
sub-MNI2_rec-uncombined15_T2starw.nii.gz: 53%|▌| 356k/667k [00:00<00:00, 1.43MB
sub-MNI2_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/816k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined16_T2starw.nii.gz: 46%|▍| 356k/780k [00:00<00:00, 1.42MB
sub-MNI2_rec-uncombined18_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined17_T2starw.nii.gz: 10%| | 83.6k/816k [00:00<00:01, 602kB
sub-MNI2_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/788k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined19_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined17_T2starw.nii.gz: 37%|▎| 305k/816k [00:00<00:00, 1.24MB
sub-MNI2_rec-uncombined18_T2starw.nii.gz: 11%| | 83.6k/788k [00:00<00:01, 612kB
sub-MNI2_acq-anat_TB1TFL.json: 0%| | 0.00/2.59k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined17_T2starw.nii.gz: 69%|▋| 561k/816k [00:00<00:00, 1.68MB
sub-MNI2_rec-uncombined18_T2starw.nii.gz: 32%|▎| 254k/788k [00:00<00:00, 1.17MB
sub-MNI2_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/666k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined20_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined18_T2starw.nii.gz: 93%|▉| 730k/788k [00:00<00:00, 2.52MB
sub-MNI2_rec-uncombined19_T2starw.nii.gz: 13%|▏| 83.6k/666k [00:00<00:01, 590kB
sub-MNI2_rec-uncombined19_T2starw.nii.gz: 53%|▌| 356k/666k [00:00<00:00, 1.53MB
sub-MNI2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/802k [00:00<?, ?B/s]
sub-MNI2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/815k [00:00<?, ?B/s]
sub-MNI2_rec-uncombined20_T2starw.nii.gz: 10%| | 83.5k/802k [00:00<00:01, 640kB
sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.13M [00:00<?, ?B/s]
sub-MNI2_acq-anat_TB1TFL.nii.gz: 10%|▋ | 83.6k/815k [00:00<00:01, 612kB/s]
sub-MNI2_rec-uncombined20_T2starw.nii.gz: 51%|▌| 407k/802k [00:00<00:00, 1.67MB
sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.5k/4.13M [00:00<00:07, 593kB/
sub-MNI2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-MNI2_acq-anat_TB1TFL.nii.gz: 25%|█▉ | 203k/815k [00:00<00:00, 898kB/s]
sub-MNI2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.65M [00:00<?, ?B/s]
sub-MNI2_acq-anat_TB1TFL.nii.gz: 85%|█████▉ | 696k/815k [00:00<00:00, 2.55MB/s]
sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz: 10%| | 407k/4.13M [00:00<00:02, 1.65MB/
sub-MNI2_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.65M [00:00<00:06, 58
sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz: 40%|▍| 1.64M/4.13M [00:00<00:00, 4.98MB
sub-MNI2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaSagSmall_GFactor.nii.gz: 9%| | 339k/3.65M [00:00<00:02, 1.2
sub-MNI2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.9M [00:00<?, ?B/s]
sub-MNI2_acq-coilQaSagSmall_GFactor.nii.gz: 39%|▍| 1.41M/3.65M [00:00<00:00, 4.
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/15.9M [00:00<00:29, 570kB/s
sub-MNI2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/91.6k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/15.9M [00:00<00:09, 1.80MB/s
sub-MNI2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.2k [00:00<?, ?B/s]
sub-MNI2_acq-famp-0.66_TB1DREAM.nii.gz: 91%|▉| 83.6k/91.6k [00:00<00:00, 591kB/
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 6%| | 986k/15.9M [00:00<00:04, 3.39MB/s
sub-MNI2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-MNI2_acq-famp-1.5_TB1DREAM.nii.gz: 92%|▉| 83.5k/91.2k [00:00<00:00, 557kB/s
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 14%|▏| 2.30M/15.9M [00:00<00:01, 7.21MB/
sub-MNI2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/91.6k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 45%|▍| 7.22M/15.9M [00:00<00:00, 21.8MB/
sub-MNI2_acq-famp_TB1DREAM.nii.gz: 93%|███▋| 85.2k/91.6k [00:00<00:00, 668kB/s]
sub-MNI2_acq-famp_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 68%|▋| 10.8M/15.9M [00:00<00:00, 25.1MB/
sub-MNI2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MNI2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/908k [00:00<?, ?B/s]
sub-MNI2_acq-coilQaTra_GFactor.nii.gz: 91%|▉| 14.5M/15.9M [00:00<00:00, 29.2MB/
sub-MNI2_acq-famp_TB1TFL.nii.gz: 9%|▋ | 83.5k/908k [00:00<00:01, 614kB/s]
sub-MNI2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-MNI2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/79.3k [00:00<?, ?B/s]
sub-MNI2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-MNI2_acq-famp_TB1TFL.nii.gz: 22%|█▊ | 203k/908k [00:00<00:00, 862kB/s]
sub-MNI2_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 79.3k/79.3k [00:00<00:00, 599kB/
sub-MNI2_acq-famp_TB1TFL.nii.gz: 80%|█████▌ | 730k/908k [00:00<00:00, 2.66MB/s]
sub-MNI2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-MNI2_acq-refv-1.5_TB1DREAM.nii.gz: 74%|▋| 66.5k/89.8k [00:00<00:00, 582kB/s
sub-MNI2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/86.5k [00:00<?, ?B/s]
sub-MNI3_T2starw.json: 0%| | 0.00/2.53k [00:00<?, ?B/s]
sub-MNI3_T2starw.nii.gz: 0%| | 0.00/1.36M [00:00<?, ?B/s]
sub-MNI2_acq-refv_TB1DREAM.nii.gz: 97%|███▊| 83.6k/86.5k [00:00<00:00, 651kB/s]
sub-MNI3_T2starw.nii.gz: 6%|▊ | 83.6k/1.36M [00:00<00:02, 648kB/s]
sub-MNI3_UNIT1.nii.gz: 0%| | 0.00/26.1M [00:00<?, ?B/s]
sub-MNI3_UNIT1.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MNI3_inv-1_MP2RAGE.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MNI3_UNIT1.nii.gz: 0%| | 50.6k/26.1M [00:00<00:57, 472kB/s]
sub-MNI3_T2starw.nii.gz: 16%|██▎ | 220k/1.36M [00:00<00:01, 936kB/s]
sub-MNI3_inv-2_MP2RAGE.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MNI3_T2starw.nii.gz: 55%|███████▋ | 765k/1.36M [00:00<00:00, 2.76MB/s]
sub-MNI3_UNIT1.nii.gz: 1%| | 169k/26.1M [00:00<00:31, 858kB/s]
sub-MNI3_UNIT1.nii.gz: 3%|▍ | 747k/26.1M [00:00<00:09, 2.91MB/s]
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.6M [00:00<?, ?B/s]
sub-MNI3_UNIT1.nii.gz: 6%|▉ | 1.54M/26.1M [00:00<00:05, 4.92MB/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.3M [00:00<?, ?B/s]
sub-MNI3_rec-uncombined01_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.5k/19.6M [00:00<00:31, 649kB/
sub-MNI3_UNIT1.nii.gz: 16%|██▍ | 4.26M/26.1M [00:00<00:01, 13.0MB/s]
sub-MNI3_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/814k [00:00<?, ?B/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.5k/17.3M [00:00<00:29, 609kB/
sub-MNI3_UNIT1.nii.gz: 35%|█████▏ | 9.06M/26.1M [00:00<00:00, 25.2MB/s]
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 407k/19.6M [00:00<00:11, 1.74MB/
sub-MNI3_UNIT1.nii.gz: 48%|███████▏ | 12.6M/26.1M [00:00<00:00, 29.0MB/s]
sub-MNI3_rec-uncombined01_T2starw.nii.gz: 10%| | 85.2k/814k [00:00<00:01, 664kB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 407k/17.3M [00:00<00:10, 1.64MB/
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.24M/19.6M [00:00<00:04, 4.50MB
sub-MNI3_rec-uncombined02_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined01_T2starw.nii.gz: 42%|▍| 340k/814k [00:00<00:00, 1.67MB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 6%| | 1.00M/17.3M [00:00<00:05, 3.38MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 10%| | 1.96M/19.6M [00:00<00:03, 5.56MB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 10%| | 1.66M/17.3M [00:00<00:03, 4.63MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 2.91M/19.6M [00:00<00:02, 7.00MB
sub-MNI3_UNIT1.nii.gz: 59%|████████▊ | 15.4M/26.1M [00:00<00:00, 20.6MB/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 17%|▏| 3.01M/17.3M [00:00<00:01, 7.77MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.77M/19.6M [00:00<00:01, 11.0MB
sub-MNI3_rec-uncombined03_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_UNIT1.nii.gz: 68%|██████████▏ | 17.7M/26.1M [00:01<00:00, 19.0MB/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.19M/17.3M [00:00<00:01, 9.25MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 31%|▎| 5.97M/19.6M [00:00<00:01, 11.5MB
sub-MNI3_UNIT1.nii.gz: 76%|███████████▍ | 19.8M/26.1M [00:01<00:00, 18.7MB/s]
sub-MNI3_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/765k [00:00<?, ?B/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 35%|▎| 6.05M/17.3M [00:00<00:00, 12.4MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 40%|▍| 7.75M/19.6M [00:00<00:00, 13.7MB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 45%|▍| 7.76M/17.3M [00:00<00:00, 14.1MB
sub-MNI3_UNIT1.nii.gz: 83%|████████████▍ | 21.8M/26.1M [00:01<00:00, 18.6MB/s]
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 49%|▍| 9.58M/19.6M [00:00<00:00, 15.3MB
sub-MNI3_rec-uncombined02_T2starw.nii.gz: 11%| | 83.6k/765k [00:00<00:01, 654kB
sub-MNI3_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/828k [00:00<?, ?B/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 55%|▌| 9.54M/17.3M [00:00<00:00, 15.5MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 58%|▌| 11.3M/19.6M [00:01<00:00, 16.1MB
sub-MNI3_UNIT1.nii.gz: 91%|█████████████▌ | 23.7M/26.1M [00:01<00:00, 18.3MB/s]
sub-MNI3_rec-uncombined02_T2starw.nii.gz: 46%|▍| 356k/765k [00:00<00:00, 1.51MB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 64%|▋| 11.2M/17.3M [00:01<00:00, 16.0MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 66%|▋| 12.9M/19.6M [00:01<00:00, 16.3MB
sub-MNI3_rec-uncombined03_T2starw.nii.gz: 10%| | 83.6k/828k [00:00<00:01, 651kB
sub-MNI3_UNIT1.nii.gz: 98%|██████████████▋| 25.5M/26.1M [00:01<00:00, 16.9MB/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 73%|▋| 12.7M/17.3M [00:01<00:00, 14.3MB
sub-MNI3_rec-uncombined03_T2starw.nii.gz: 43%|▍| 356k/828k [00:00<00:00, 1.52MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 74%|▋| 14.5M/19.6M [00:01<00:00, 14.7MB
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 83%|▊| 14.4M/17.3M [00:01<00:00, 15.2MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 82%|▊| 16.1M/19.6M [00:01<00:00, 15.4MB
sub-MNI3_rec-uncombined04_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz: 95%|▉| 16.4M/17.3M [00:01<00:00, 16.8MB
sub-MNI3_inv-2_part-mag_MP2RAGE.nii.gz: 93%|▉| 18.1M/19.6M [00:01<00:00, 17.1MB
sub-MNI3_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/898k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined04_T2starw.nii.gz: 9%| | 85.2k/898k [00:00<00:01, 692kB
sub-MNI3_rec-uncombined05_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined06_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/844k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined04_T2starw.nii.gz: 40%|▍| 357k/898k [00:00<00:00, 1.49MB
sub-MNI3_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/821k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined05_T2starw.nii.gz: 10%| | 83.6k/844k [00:00<00:01, 651kB
sub-MNI3_rec-uncombined07_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined06_T2starw.nii.gz: 10%| | 83.6k/821k [00:00<00:01, 692kB
sub-MNI3_rec-uncombined05_T2starw.nii.gz: 42%|▍| 356k/844k [00:00<00:00, 1.63MB
sub-MNI3_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/872k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined06_T2starw.nii.gz: 31%|▎| 254k/821k [00:00<00:00, 1.13MB
sub-MNI3_rec-uncombined07_T2starw.nii.gz: 10%| | 83.6k/872k [00:00<00:01, 612kB
sub-MNI3_rec-uncombined09_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined06_T2starw.nii.gz: 87%|▊| 713k/821k [00:00<00:00, 2.56MB
sub-MNI3_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/945k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined07_T2starw.nii.gz: 41%|▍| 356k/872k [00:00<00:00, 1.38MB
sub-MNI3_rec-uncombined08_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined08_T2starw.nii.gz: 9%| | 83.6k/945k [00:00<00:01, 603kB
sub-MNI3_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/847k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/861k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined08_T2starw.nii.gz: 38%|▍| 356k/945k [00:00<00:00, 1.53MB
sub-MNI3_rec-uncombined09_T2starw.nii.gz: 10%| | 83.6k/861k [00:00<00:01, 648kB
sub-MNI3_rec-uncombined10_T2starw.nii.gz: 11%| | 94.0k/847k [00:00<00:01, 683kB
sub-MNI3_rec-uncombined10_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/903k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined09_T2starw.nii.gz: 41%|▍| 356k/861k [00:00<00:00, 1.51MB
sub-MNI3_rec-uncombined10_T2starw.nii.gz: 47%|▍| 400k/847k [00:00<00:00, 1.59MB
sub-MNI3_rec-uncombined11_T2starw.nii.gz: 9%| | 85.2k/903k [00:00<00:01, 667kB
sub-MNI3_rec-uncombined11_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined10_T2starw.nii.gz: 100%|█| 847k/847k [00:00<00:00, 2.63MB
sub-MNI3_rec-uncombined12_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined11_T2starw.nii.gz: 40%|▍| 357k/903k [00:00<00:00, 1.46MB
sub-MNI3_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/933k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined13_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined14_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/806k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined12_T2starw.nii.gz: 9%| | 83.6k/933k [00:00<00:01, 656kB
sub-MNI3_rec-uncombined15_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined13_T2starw.nii.gz: 11%| | 85.2k/806k [00:00<00:01, 656kB
sub-MNI3_rec-uncombined12_T2starw.nii.gz: 38%|▍| 356k/933k [00:00<00:00, 1.52MB
sub-MNI3_rec-uncombined13_T2starw.nii.gz: 44%|▍| 357k/806k [00:00<00:00, 1.39MB
sub-MNI3_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/815k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/966k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/750k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined14_T2starw.nii.gz: 10%| | 83.6k/815k [00:00<00:01, 557kB
sub-MNI3_rec-uncombined16_T2starw.nii.gz: 8%| | 77.0k/966k [00:00<00:01, 564kB
sub-MNI3_rec-uncombined16_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined14_T2starw.nii.gz: 44%|▍| 356k/815k [00:00<00:00, 1.36MB
sub-MNI3_rec-uncombined15_T2starw.nii.gz: 11%| | 83.6k/750k [00:00<00:01, 598kB
sub-MNI3_rec-uncombined16_T2starw.nii.gz: 38%|▍| 366k/966k [00:00<00:00, 1.47MB
sub-MNI3_rec-uncombined17_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined15_T2starw.nii.gz: 38%|▍| 288k/750k [00:00<00:00, 1.27MB
sub-MNI3_rec-uncombined16_T2starw.nii.gz: 100%|▉| 962k/966k [00:00<00:00, 3.20MB
sub-MNI3_rec-uncombined18_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-MNI3_rec-uncombined19_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/965k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/1.02M [00:00<00:01, 654k
sub-MNI3_rec-uncombined20_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined18_T2starw.nii.gz: 9%| | 83.6k/965k [00:00<00:01, 649kB
sub-MNI3_rec-uncombined17_T2starw.nii.gz: 34%|▎| 356k/1.02M [00:00<00:00, 1.51M
sub-MNI3_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/912k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined18_T2starw.nii.gz: 37%|▎| 356k/965k [00:00<00:00, 1.51MB
sub-MNI3_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/959k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined19_T2starw.nii.gz: 9%| | 83.6k/912k [00:00<00:01, 651kB
sub-MNI3_rec-uncombined20_T2starw.nii.gz: 9%| | 85.2k/959k [00:00<00:01, 769kB
sub-MNI3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/921k [00:00<?, ?B/s]
sub-MNI3_rec-uncombined19_T2starw.nii.gz: 39%|▍| 356k/912k [00:00<00:00, 1.52MB
sub-MNI3_rec-uncombined20_T2starw.nii.gz: 37%|▎| 357k/959k [00:00<00:00, 1.60MB
sub-MNI3_acq-anat_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-MNI3_acq-anat_TB1TFL.nii.gz: 9%|▋ | 83.6k/921k [00:00<00:01, 560kB/s]
sub-MNI3_acq-anat_TB1TFL.nii.gz: 18%|█▍ | 169k/921k [00:00<00:01, 670kB/s]
sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.52M [00:00<?, ?B/s]
sub-MNI3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.23k [00:00<?, ?B/s]
sub-MNI3_acq-anat_TB1TFL.nii.gz: 31%|██▍ | 288k/921k [00:00<00:00, 731kB/s]
sub-MNI3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.25k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.52M [00:00<00:08, 531kB/
sub-MNI3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.65M [00:00<?, ?B/s]
sub-MNI3_acq-anat_TB1TFL.nii.gz: 44%|███▌ | 407k/921k [00:00<00:00, 825kB/s]
sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.52M [00:00<00:02, 1.46MB/
sub-MNI3_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.65M [00:00<00:05, 65
sub-MNI3_acq-anat_TB1TFL.nii.gz: 57%|████▌ | 527k/921k [00:00<00:00, 881kB/s]
sub-MNI3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz: 32%|▎| 1.46M/4.52M [00:00<00:00, 4.47MB
sub-MNI3_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.65M [00:00<00:02, 1.5
sub-MNI3_acq-anat_TB1TFL.nii.gz: 74%|█████▉ | 679k/921k [00:00<00:00, 983kB/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.1M [00:00<?, ?B/s]
sub-MNI3_acq-coilQaSagSmall_GFactor.nii.gz: 38%|▍| 1.39M/3.65M [00:00<00:00, 5.
sub-MNI3_acq-anat_TB1TFL.nii.gz: 90%|██████▎| 833k/921k [00:00<00:00, 1.05MB/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 0%| | 77.0k/16.1M [00:00<00:29, 569kB/s
sub-MNI3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaSagSmall_GFactor.nii.gz: 70%|▋| 2.57M/3.65M [00:00<00:00, 7.
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 2%| | 366k/16.1M [00:00<00:11, 1.48MB/s
sub-MNI3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.8k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 9%| | 1.45M/16.1M [00:00<00:03, 4.57MB/
sub-MNI3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-MNI3_acq-famp_TB1TFL.json: 0%| | 0.00/2.74k [00:00<?, ?B/s]
sub-MNI3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 22%|▏| 3.58M/16.1M [00:00<00:01, 9.94MB/
sub-MNI3_acq-famp-0.66_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.8k [00:00<00:00, 555kB/
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 45%|▍| 7.30M/16.1M [00:00<00:00, 18.8MB/
sub-MNI3_acq-famp_TB1DREAM.nii.gz: 91%|███▋| 83.6k/92.1k [00:00<00:00, 609kB/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 69%|▋| 11.1M/16.1M [00:00<00:00, 25.2MB/
sub-MNI3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/946k [00:00<?, ?B/s]
sub-MNI3_acq-coilQaTra_GFactor.nii.gz: 98%|▉| 15.9M/16.1M [00:00<00:00, 32.8MB/
sub-MNI3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.22k [00:00<?, ?B/s]
sub-MNI3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.2k [00:00<?, ?B/s]
sub-MNI3_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/946k [00:00<00:01, 617kB/s]
sub-MNI3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-MNI3_acq-refv-0.66_TB1DREAM.nii.gz: 97%|▉| 85.2k/88.2k [00:00<00:00, 732kB/
sub-MNI3_acq-famp_TB1TFL.nii.gz: 41%|██▉ | 390k/946k [00:00<00:00, 1.58MB/s]
sub-MPI1_T2starw.json: 0%| | 0.00/2.38k [00:00<?, ?B/s]
sub-MNI3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/93.7k [00:00<?, ?B/s]
sub-MPI1_UNIT1.json: 0%| | 0.00/2.04k [00:00<?, ?B/s]
sub-MNI3_acq-refv_TB1DREAM.nii.gz: 89%|███▌| 83.6k/93.7k [00:00<00:00, 778kB/s]
sub-MPI1_T2starw.nii.gz: 0%| | 0.00/1.29M [00:00<?, ?B/s]
sub-MPI1_T2starw.nii.gz: 5%|▋ | 67.6k/1.29M [00:00<00:02, 578kB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.5M [00:00<?, ?B/s]
sub-MPI1_inv-1_MP2RAGE.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI1_UNIT1.nii.gz: 0%| | 0.00/25.0M [00:00<?, ?B/s]
sub-MPI1_inv-2_MP2RAGE.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI1_T2starw.nii.gz: 24%|███▍ | 322k/1.29M [00:00<00:00, 1.38MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/18.5M [00:00<00:30, 627kB/
sub-MPI1_UNIT1.nii.gz: 0%| | 83.6k/25.0M [00:00<00:35, 746kB/s]
sub-MPI1_T2starw.nii.gz: 100%|█████████████| 1.29M/1.29M [00:00<00:00, 4.67MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 357k/18.5M [00:00<00:13, 1.43MB/
sub-MPI1_UNIT1.nii.gz: 2%|▏ | 390k/25.0M [00:00<00:14, 1.77MB/s]
sub-MPI1_rec-uncombined10_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.8M [00:00<?, ?B/s]
sub-MPI1_UNIT1.nii.gz: 5%|▋ | 1.16M/25.0M [00:00<00:05, 4.35MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.23M/18.5M [00:00<00:04, 4.22MB
sub-MPI1_UNIT1.nii.gz: 12%|█▊ | 3.12M/25.0M [00:00<00:02, 10.3MB/s]
sub-MPI1_rec-uncombined11_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 2.93M/18.5M [00:00<00:02, 8.14MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/19.8M [00:00<00:33, 618kB/
sub-MPI1_UNIT1.nii.gz: 20%|███ | 5.04M/25.0M [00:00<00:01, 13.6MB/s]
sub-MPI1_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/980k [00:00<?, ?B/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 28%|▎| 5.26M/18.5M [00:00<00:01, 13.2MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 373k/19.8M [00:00<00:12, 1.60MB/
sub-MPI1_UNIT1.nii.gz: 29%|████▎ | 7.25M/25.0M [00:00<00:01, 16.7MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 41%|▍| 7.49M/18.5M [00:00<00:00, 16.3MB
sub-MPI1_rec-uncombined10_T2starw.nii.gz: 9%| | 85.2k/980k [00:00<00:01, 655kB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.09M/19.8M [00:00<00:04, 3.95MB
sub-MPI1_UNIT1.nii.gz: 37%|█████▌ | 9.23M/25.0M [00:00<00:00, 18.0MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 50%|▍| 9.23M/18.5M [00:00<00:00, 16.8MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.56M/19.8M [00:00<00:02, 8.02MB
sub-MPI1_rec-uncombined10_T2starw.nii.gz: 36%|▎| 357k/980k [00:00<00:00, 1.51MB
sub-MPI1_UNIT1.nii.gz: 44%|██████▌ | 11.0M/25.0M [00:00<00:00, 17.9MB/s]
sub-MPI1_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/977k [00:00<?, ?B/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 59%|▌| 10.9M/18.5M [00:00<00:00, 17.1MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 21%|▏| 4.18M/19.8M [00:00<00:01, 11.1MB
sub-MPI1_rec-uncombined10_T2starw.nii.gz: 100%|█| 980k/980k [00:00<00:00, 3.39MB
sub-MPI1_UNIT1.nii.gz: 51%|███████▋ | 12.7M/25.0M [00:00<00:00, 16.7MB/s]
sub-MPI1_rec-uncombined11_T2starw.nii.gz: 9%| | 83.6k/977k [00:00<00:01, 657kB
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 68%|▋| 12.6M/18.5M [00:01<00:00, 16.1MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 27%|▎| 5.43M/19.8M [00:00<00:01, 11.7MB
sub-MPI1_UNIT1.nii.gz: 58%|████████▋ | 14.5M/25.0M [00:01<00:00, 17.1MB/s]
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 78%|▊| 14.4M/18.5M [00:01<00:00, 16.9MB
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 37%|▎| 7.23M/19.8M [00:00<00:00, 14.0MB
sub-MPI1_rec-uncombined11_T2starw.nii.gz: 36%|▎| 356k/977k [00:00<00:00, 1.52MB
sub-MPI1_UNIT1.nii.gz: 65%|█████████▋ | 16.2M/25.0M [00:01<00:00, 17.3MB/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 45%|▍| 8.84M/19.8M [00:00<00:00, 14.8MB
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 87%|▊| 16.1M/18.5M [00:01<00:00, 16.8MB
sub-MPI1_rec-uncombined12_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_UNIT1.nii.gz: 71%|██████████▋ | 17.9M/25.0M [00:01<00:00, 15.5MB/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 10.3M/19.8M [00:00<00:00, 14.0MB
sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz: 96%|▉| 17.7M/18.5M [00:01<00:00, 15.4MB
sub-MPI1_UNIT1.nii.gz: 78%|███████████▋ | 19.4M/25.0M [00:01<00:00, 15.5MB/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 60%|▌| 11.8M/19.8M [00:01<00:00, 14.6MB
sub-MPI1_UNIT1.nii.gz: 89%|█████████████▍ | 22.4M/25.0M [00:01<00:00, 19.9MB/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 75%|▋| 14.8M/19.8M [00:01<00:00, 19.5MB
sub-MPI1_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/969k [00:00<?, ?B/s]
sub-MPI1_inv-2_part-mag_MP2RAGE.nii.gz: 88%|▉| 17.4M/19.8M [00:01<00:00, 20.6MB
sub-MPI1_rec-uncombined13_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/954k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined12_T2starw.nii.gz: 7%| | 66.6k/969k [00:00<00:01, 619kB
sub-MPI1_rec-uncombined13_T2starw.nii.gz: 9%| | 85.2k/954k [00:00<00:01, 722kB
sub-MPI1_rec-uncombined12_T2starw.nii.gz: 23%|▏| 221k/969k [00:00<00:00, 1.16MB
sub-MPI1_rec-uncombined14_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined13_T2starw.nii.gz: 37%|▎| 357k/954k [00:00<00:00, 1.56MB
sub-MPI1_rec-uncombined12_T2starw.nii.gz: 75%|▊| 730k/969k [00:00<00:00, 2.41MB
sub-MPI1_rec-uncombined15_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined14_T2starw.nii.gz: 8%| | 85.2k/1.03M [00:00<00:01, 657k
sub-MPI1_rec-uncombined16_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined17_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/971k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined14_T2starw.nii.gz: 34%|▎| 357k/1.03M [00:00<00:00, 1.50M
sub-MPI1_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined15_T2starw.nii.gz: 8%| | 77.0k/971k [00:00<00:01, 526kB
sub-MPI1_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 673k
sub-MPI1_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined15_T2starw.nii.gz: 38%|▍| 366k/971k [00:00<00:00, 1.43MB
sub-MPI1_rec-uncombined18_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined16_T2starw.nii.gz: 34%|▎| 356k/1.01M [00:00<00:00, 1.47M
sub-MPI1_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 615k
sub-MPI1_rec-uncombined19_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined16_T2starw.nii.gz: 92%|▉| 951k/1.01M [00:00<00:00, 3.17M
sub-MPI1_rec-uncombined17_T2starw.nii.gz: 28%|▎| 288k/1.01M [00:00<00:00, 1.35M
sub-MPI1_rec-uncombined17_T2starw.nii.gz: 75%|▊| 781k/1.01M [00:00<00:00, 2.79M
sub-MPI1_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined1_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined18_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 664k
sub-MPI1_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined20_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined18_T2starw.nii.gz: 33%|▎| 356k/1.04M [00:00<00:00, 1.51M
sub-MPI1_rec-uncombined19_T2starw.nii.gz: 8%| | 83.6k/1.06M [00:00<00:01, 650k
sub-MPI1_rec-uncombined1_T2starw.nii.gz: 8%| | 85.2k/0.99M [00:00<00:01, 592kB
sub-MPI1_rec-uncombined21_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined19_T2starw.nii.gz: 33%|▎| 356k/1.06M [00:00<00:00, 1.51M
sub-MPI1_rec-uncombined1_T2starw.nii.gz: 35%|▎| 357k/0.99M [00:00<00:00, 1.31MB
sub-MPI1_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined22_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 651k
sub-MPI1_rec-uncombined23_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/1.03M [00:00<00:01, 781k
sub-MPI1_rec-uncombined20_T2starw.nii.gz: 33%|▎| 356k/1.05M [00:00<00:00, 1.51M
sub-MPI1_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined21_T2starw.nii.gz: 34%|▎| 356k/1.03M [00:00<00:00, 1.63M
sub-MPI1_rec-uncombined23_T2starw.nii.gz: 6%| | 67.6k/1.04M [00:00<00:01, 530k
sub-MPI1_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined24_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined23_T2starw.nii.gz: 32%|▎| 339k/1.04M [00:00<00:00, 1.50M
sub-MPI1_rec-uncombined22_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 608k
sub-MPI1_rec-uncombined2_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined22_T2starw.nii.gz: 19%|▏| 203k/1.05M [00:00<00:01, 873kB
sub-MPI1_rec-uncombined3_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined22_T2starw.nii.gz: 66%|▋| 713k/1.05M [00:00<00:00, 2.48M
sub-MPI1_rec-uncombined24_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 794k
sub-MPI1_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/993k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined24_T2starw.nii.gz: 35%|▎| 356k/0.99M [00:00<00:00, 1.63M
sub-MPI1_rec-uncombined4_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined2_T2starw.nii.gz: 9%| | 85.2k/968k [00:00<00:01, 664kB/
sub-MPI1_rec-uncombined3_T2starw.nii.gz: 8%| | 83.6k/993k [00:00<00:01, 614kB/
sub-MPI1_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/967k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined2_T2starw.nii.gz: 39%|▍| 375k/968k [00:00<00:00, 1.60MB/
sub-MPI1_rec-uncombined3_T2starw.nii.gz: 36%|▎| 356k/993k [00:00<00:00, 1.42MB/
sub-MPI1_rec-uncombined4_T2starw.nii.gz: 9%| | 83.6k/967k [00:00<00:01, 556kB/
sub-MPI1_rec-uncombined5_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined6_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined4_T2starw.nii.gz: 37%|▎| 356k/967k [00:00<00:00, 1.54MB/
sub-MPI1_rec-uncombined7_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/967k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined8_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined5_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 638kB
sub-MPI1_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI1_rec-uncombined6_T2starw.nii.gz: 9%| | 83.6k/967k [00:00<00:01, 616kB/
sub-MPI1_rec-uncombined5_T2starw.nii.gz: 35%|▎| 356k/0.98M [00:00<00:00, 1.44MB
sub-MPI1_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/996k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined7_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 616kB
sub-MPI1_rec-uncombined6_T2starw.nii.gz: 37%|▎| 356k/967k [00:00<00:00, 1.43MB/
sub-MPI1_rec-uncombined9_T2starw.nii.gz: 0%| | 0.00/981k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined8_T2starw.nii.gz: 9%| | 85.2k/996k [00:00<00:01, 625kB/
sub-MPI1_rec-uncombined7_T2starw.nii.gz: 34%|▎| 356k/1.01M [00:00<00:00, 1.43MB
sub-MPI1_rec-uncombined9_T2starw.nii.gz: 9%| | 83.6k/981k [00:00<00:01, 540kB/
sub-MPI1_rec-uncombined8_T2starw.nii.gz: 34%|▎| 340k/996k [00:00<00:00, 1.40MB/
sub-MPI1_acq-anat_TB1TFL.json: 0%| | 0.00/2.62k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined9_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI1_rec-uncombined9_T2starw.nii.gz: 35%|▎| 339k/981k [00:00<00:00, 1.26MB/
sub-MPI1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MPI1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/844k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.99M [00:00<?, ?B/s]
sub-MPI1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI1_acq-anat_TB1TFL.nii.gz: 10%|▋ | 85.2k/844k [00:00<00:01, 618kB/s]
sub-MPI1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.59M [00:00<?, ?B/s]
sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.99M [00:00<00:06, 597kB/
sub-MPI1_acq-anat_TB1TFL.nii.gz: 42%|██▉ | 357k/844k [00:00<00:00, 1.53MB/s]
sub-MPI1_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.59M [00:00<00:05, 65
sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 356k/3.99M [00:00<00:02, 1.38MB/
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.6M [00:00<?, ?B/s]
sub-MPI1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.59M [00:00<00:02, 1.4
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/15.6M [00:00<00:24, 668kB/s
sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz: 37%|▎| 1.46M/3.99M [00:00<00:00, 4.55MB
sub-MPI1_acq-coilQaSagSmall_GFactor.nii.gz: 41%|▍| 1.46M/3.59M [00:00<00:00, 4.
sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz: 80%|▊| 3.20M/3.99M [00:00<00:00, 8.90MB
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/15.6M [00:00<00:10, 1.52MB/s
sub-MPI1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 5%| | 782k/15.6M [00:00<00:06, 2.57MB/s
sub-MPI1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.2k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 18%|▏| 2.85M/15.6M [00:00<00:01, 9.38MB/
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 46%|▍| 7.17M/15.6M [00:00<00:00, 21.4MB/
sub-MPI1_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI1_acq-famp-0.66_TB1DREAM.nii.gz: 96%|▉| 83.5k/87.2k [00:00<00:00, 524kB/
sub-MPI1_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/88.6k [00:00<?, ?B/s]
sub-MPI1_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 60%|▌| 9.35M/15.6M [00:00<00:00, 20.4MB/
sub-MPI1_acq-famp-1.5_TB1DREAM.nii.gz: 96%|▉| 85.2k/88.6k [00:00<00:00, 654kB/s
sub-MPI1_acq-coilQaTra_GFactor.nii.gz: 87%|▊| 13.5M/15.6M [00:00<00:00, 26.8MB/
sub-MPI1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/87.9k [00:00<?, ?B/s]
sub-MPI1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/82.7k [00:00<?, ?B/s]
sub-MPI1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/948k [00:00<?, ?B/s]
sub-MPI1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI1_acq-famp_TB1DREAM.nii.gz: 100%|████| 87.9k/87.9k [00:00<00:00, 642kB/s]
sub-MPI1_acq-famp_TB1TFL.nii.gz: 5%|▎ | 49.6k/948k [00:00<00:01, 477kB/s]
sub-MPI1_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 82.7k/82.7k [00:00<00:00, 554kB/
sub-MPI1_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/90.7k [00:00<?, ?B/s]
sub-MPI1_acq-famp_TB1TFL.nii.gz: 25%|█▊ | 239k/948k [00:00<00:00, 1.26MB/s]
sub-MPI1_acq-refv-1.5_TB1DREAM.nii.gz: 92%|▉| 83.5k/90.7k [00:00<00:00, 616kB/s
sub-MPI1_acq-famp_TB1TFL.nii.gz: 77%|█████▍ | 731k/948k [00:00<00:00, 2.53MB/s]
sub-MPI1_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI2_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI2_T2starw.nii.gz: 0%| | 0.00/1.27M [00:00<?, ?B/s]
sub-MPI2_UNIT1.json: 0%| | 0.00/2.03k [00:00<?, ?B/s]
sub-MPI1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/88.0k [00:00<?, ?B/s]
sub-MPI2_inv-1_MP2RAGE.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI2_UNIT1.nii.gz: 0%| | 0.00/25.5M [00:00<?, ?B/s]
sub-MPI2_T2starw.nii.gz: 6%|▉ | 83.6k/1.27M [00:00<00:01, 646kB/s]
sub-MPI1_acq-refv_TB1DREAM.nii.gz: 88%|███▌| 77.0k/88.0k [00:00<00:00, 587kB/s]
sub-MPI2_T2starw.nii.gz: 26%|███▋ | 340k/1.27M [00:00<00:00, 1.42MB/s]
sub-MPI2_UNIT1.nii.gz: 0%| | 83.6k/25.5M [00:00<00:43, 607kB/s]
sub-MPI2_inv-2_MP2RAGE.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI2_T2starw.nii.gz: 64%|████████▉ | 832k/1.27M [00:00<00:00, 2.82MB/s]
sub-MPI2_UNIT1.nii.gz: 1%|▏ | 323k/25.5M [00:00<00:17, 1.53MB/s]
sub-MPI2_UNIT1.nii.gz: 3%|▍ | 730k/25.5M [00:00<00:10, 2.51MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.2M [00:00<?, ?B/s]
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.4M [00:00<?, ?B/s]
sub-MPI2_UNIT1.nii.gz: 10%|█▌ | 2.55M/25.5M [00:00<00:02, 8.58MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 94.0k/18.2M [00:00<00:27, 685kB/
sub-MPI2_rec-uncombined10_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_UNIT1.nii.gz: 18%|██▋ | 4.47M/25.5M [00:00<00:01, 12.5MB/s]
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/19.4M [00:00<00:32, 622kB/
sub-MPI2_rec-uncombined11_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_UNIT1.nii.gz: 30%|████▍ | 7.61M/25.5M [00:00<00:00, 19.2MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 400k/18.2M [00:00<00:11, 1.59MB/
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 408k/19.4M [00:00<00:11, 1.78MB/
sub-MPI2_UNIT1.nii.gz: 41%|██████▏ | 10.5M/25.5M [00:00<00:00, 22.8MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.19M/18.2M [00:00<00:04, 4.11MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.18M/19.4M [00:00<00:04, 4.29MB
sub-MPI2_rec-uncombined12_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 10%| | 1.75M/18.2M [00:00<00:03, 4.74MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 11%| | 2.08M/19.4M [00:00<00:02, 6.11MB
sub-MPI2_UNIT1.nii.gz: 50%|███████▌ | 12.8M/25.5M [00:00<00:00, 19.9MB/s]
sub-MPI2_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 18%|▏| 3.22M/18.2M [00:00<00:01, 8.27MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 20%|▏| 3.85M/19.4M [00:00<00:01, 10.3MB
sub-MPI2_UNIT1.nii.gz: 58%|████████▋ | 14.8M/25.5M [00:01<00:00, 19.5MB/s]
sub-MPI2_rec-uncombined10_T2starw.nii.gz: 8%| | 77.0k/0.98M [00:00<00:01, 600k
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 27%|▎| 4.98M/18.2M [00:00<00:01, 11.6MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 29%|▎| 5.63M/19.4M [00:00<00:01, 13.0MB
sub-MPI2_UNIT1.nii.gz: 66%|█████████▊ | 16.7M/25.5M [00:01<00:00, 19.5MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 37%|▎| 6.75M/18.2M [00:00<00:00, 13.7MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 38%|▍| 7.39M/19.4M [00:00<00:00, 14.7MB
sub-MPI2_rec-uncombined10_T2starw.nii.gz: 38%|▍| 383k/0.98M [00:00<00:00, 1.65M
sub-MPI2_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/971k [00:00<?, ?B/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 46%|▍| 8.31M/18.2M [00:00<00:00, 14.5MB
sub-MPI2_UNIT1.nii.gz: 73%|██████████▉ | 18.6M/25.5M [00:01<00:00, 17.5MB/s]
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 46%|▍| 8.89M/19.4M [00:00<00:00, 15.0MB
sub-MPI2_rec-uncombined11_T2starw.nii.gz: 9%| | 85.2k/971k [00:00<00:01, 628kB
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 53%|▌| 9.73M/18.2M [00:00<00:00, 14.4MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 53%|▌| 10.4M/19.4M [00:00<00:00, 14.9MB
sub-MPI2_UNIT1.nii.gz: 80%|███████████▉ | 20.3M/25.5M [00:01<00:00, 16.2MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 63%|▋| 11.5M/18.2M [00:01<00:00, 15.5MB
sub-MPI2_rec-uncombined11_T2starw.nii.gz: 37%|▎| 357k/971k [00:00<00:00, 1.43MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 62%|▌| 12.1M/19.4M [00:01<00:00, 15.9MB
sub-MPI2_UNIT1.nii.gz: 87%|████████████▉ | 22.0M/25.5M [00:01<00:00, 16.6MB/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 72%|▋| 13.1M/18.2M [00:01<00:00, 16.1MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 70%|▋| 13.6M/19.4M [00:01<00:00, 14.9MB
sub-MPI2_UNIT1.nii.gz: 93%|█████████████▉ | 23.7M/25.5M [00:01<00:00, 15.7MB/s]
sub-MPI2_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 81%|▊| 14.7M/18.2M [00:01<00:00, 15.6MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 80%|▊| 15.5M/19.4M [00:01<00:00, 16.3MB
sub-MPI2_rec-uncombined12_T2starw.nii.gz: 7%| | 67.5k/0.98M [00:00<00:01, 685k
sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz: 89%|▉| 16.3M/18.2M [00:01<00:00, 15.9MB
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 89%|▉| 17.3M/19.4M [00:01<00:00, 17.1MB
sub-MPI2_rec-uncombined12_T2starw.nii.gz: 17%|▏| 169k/0.98M [00:00<00:01, 776kB
sub-MPI2_rec-uncombined13_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_inv-2_part-mag_MP2RAGE.nii.gz: 100%|▉| 19.3M/19.4M [00:01<00:00, 17.5MB
sub-MPI2_rec-uncombined12_T2starw.nii.gz: 74%|▋| 747k/0.98M [00:00<00:00, 2.68M
sub-MPI2_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined14_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined13_T2starw.nii.gz: 9%| | 83.6k/968k [00:00<00:01, 649kB
sub-MPI2_rec-uncombined15_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined16_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined14_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 597k
sub-MPI2_rec-uncombined13_T2starw.nii.gz: 37%|▎| 356k/968k [00:00<00:00, 1.51MB
sub-MPI2_rec-uncombined14_T2starw.nii.gz: 32%|▎| 340k/1.05M [00:00<00:00, 1.32M
sub-MPI2_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/984k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined17_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined15_T2starw.nii.gz: 8%| | 78.0k/984k [00:00<00:01, 598kB
sub-MPI2_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/1.06M [00:00<00:01, 653k
sub-MPI2_rec-uncombined15_T2starw.nii.gz: 37%|▎| 366k/984k [00:00<00:00, 1.57MB
sub-MPI2_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined16_T2starw.nii.gz: 36%|▎| 390k/1.06M [00:00<00:00, 1.67M
sub-MPI2_rec-uncombined18_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined17_T2starw.nii.gz: 8%| | 85.2k/1.00M [00:00<00:01, 644k
sub-MPI2_rec-uncombined18_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 670k
sub-MPI2_rec-uncombined17_T2starw.nii.gz: 40%|▍| 408k/1.00M [00:00<00:00, 1.66M
sub-MPI2_rec-uncombined1_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined18_T2starw.nii.gz: 33%|▎| 356k/1.04M [00:00<00:00, 1.46M
sub-MPI2_rec-uncombined19_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined18_T2starw.nii.gz: 100%|█| 1.04M/1.04M [00:00<00:00, 3.65
sub-MPI2_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined19_T2starw.nii.gz: 7%| | 77.0k/1.03M [00:00<00:01, 586k
sub-MPI2_rec-uncombined20_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined21_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined19_T2starw.nii.gz: 35%|▎| 366k/1.03M [00:00<00:00, 1.49M
sub-MPI2_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined1_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 736kB
sub-MPI2_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 646k
sub-MPI2_rec-uncombined1_T2starw.nii.gz: 33%|▎| 339k/0.99M [00:00<00:00, 1.50MB
sub-MPI2_rec-uncombined20_T2starw.nii.gz: 33%|▎| 356k/1.04M [00:00<00:00, 1.50M
sub-MPI2_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined22_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined20_T2starw.nii.gz: 92%|▉| 985k/1.04M [00:00<00:00, 3.41M
sub-MPI2_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/1.03M [00:00<00:01, 614k
sub-MPI2_rec-uncombined22_T2starw.nii.gz: 8%| | 85.2k/1.06M [00:00<00:01, 659k
sub-MPI2_rec-uncombined21_T2starw.nii.gz: 34%|▎| 356k/1.03M [00:00<00:00, 1.42M
sub-MPI2_rec-uncombined22_T2starw.nii.gz: 33%|▎| 357k/1.06M [00:00<00:00, 1.46M
sub-MPI2_rec-uncombined24_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined23_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined22_T2starw.nii.gz: 71%|▋| 765k/1.06M [00:00<00:00, 2.41M
sub-MPI2_rec-uncombined23_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 763k
sub-MPI2_rec-uncombined23_T2starw.nii.gz: 34%|▎| 356k/1.04M [00:00<00:00, 1.61M
sub-MPI2_rec-uncombined2_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined3_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined23_T2starw.nii.gz: 94%|▉| 0.98M/1.04M [00:00<00:00, 3.46
sub-MPI2_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/959k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined24_T2starw.nii.gz: 8%| | 77.0k/0.99M [00:00<00:01, 653k
sub-MPI2_rec-uncombined2_T2starw.nii.gz: 9%| | 83.6k/959k [00:00<00:01, 693kB/
sub-MPI2_rec-uncombined4_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined24_T2starw.nii.gz: 24%|▏| 247k/0.99M [00:00<00:00, 1.24M
sub-MPI2_rec-uncombined2_T2starw.nii.gz: 30%|▎| 289k/959k [00:00<00:00, 1.45MB/
sub-MPI2_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/971k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined24_T2starw.nii.gz: 69%|▋| 706k/0.99M [00:00<00:00, 2.61M
sub-MPI2_rec-uncombined2_T2starw.nii.gz: 81%|▊| 781k/959k [00:00<00:00, 2.82MB/
sub-MPI2_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/999k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined4_T2starw.nii.gz: 9%| | 83.6k/971k [00:00<00:01, 651kB/
sub-MPI2_rec-uncombined5_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined3_T2starw.nii.gz: 8%| | 83.6k/999k [00:00<00:01, 611kB/
sub-MPI2_rec-uncombined4_T2starw.nii.gz: 37%|▎| 356k/971k [00:00<00:00, 1.51MB/
sub-MPI2_rec-uncombined6_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined3_T2starw.nii.gz: 34%|▎| 339k/999k [00:00<00:00, 1.25MB/
sub-MPI2_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/981k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined7_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined5_T2starw.nii.gz: 9%| | 83.6k/981k [00:00<00:01, 614kB/
sub-MPI2_rec-uncombined5_T2starw.nii.gz: 35%|▎| 340k/981k [00:00<00:00, 1.35MB/
sub-MPI2_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/970k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined8_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-MPI2_rec-uncombined6_T2starw.nii.gz: 10%| | 94.0k/970k [00:00<00:01, 726kB/
sub-MPI2_rec-uncombined7_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 758kB
sub-MPI2_rec-uncombined6_T2starw.nii.gz: 41%|▍| 400k/970k [00:00<00:00, 1.70MB/
sub-MPI2_rec-uncombined8_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 649kB
sub-MPI2_rec-uncombined7_T2starw.nii.gz: 35%|▎| 356k/1.00M [00:00<00:00, 1.60MB
sub-MPI2_rec-uncombined9_T2starw.json: 0%| | 0.00/2.41k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined8_T2starw.nii.gz: 34%|▎| 339k/0.98M [00:00<00:00, 1.66MB
sub-MPI2_rec-uncombined9_T2starw.nii.gz: 0%| | 0.00/994k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined7_T2starw.nii.gz: 84%|▊| 867k/1.00M [00:00<00:00, 3.06MB
sub-MPI2_rec-uncombined8_T2starw.nii.gz: 82%|▊| 816k/0.98M [00:00<00:00, 3.03MB
sub-MPI2_rec-uncombined9_T2starw.nii.gz: 8%| | 83.6k/994k [00:00<00:01, 702kB/
sub-MPI2_acq-anat_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-MPI2_rec-uncombined9_T2starw.nii.gz: 31%|▎| 305k/994k [00:00<00:00, 1.24MB/
sub-MPI2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/757k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.94M [00:00<?, ?B/s]
sub-MPI2_acq-anat_TB1TFL.nii.gz: 11%|▊ | 83.6k/757k [00:00<00:01, 624kB/s]
sub-MPI2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.94M [00:00<00:07, 575kB/
sub-MPI2_acq-anat_TB1TFL.nii.gz: 51%|███▌ | 390k/757k [00:00<00:00, 1.58MB/s]
sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 356k/3.94M [00:00<00:02, 1.50MB/
sub-MPI2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.64M [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz: 32%|▎| 1.24M/3.94M [00:00<00:00, 4.57MB
sub-MPI2_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.5k/3.64M [00:00<00:06, 59
sub-MPI2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz: 74%|▋| 2.92M/3.94M [00:00<00:00, 8.19MB
sub-MPI2_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.64M [00:00<00:02, 1.4
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.5M [00:00<?, ?B/s]
sub-MPI2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.0k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaSagSmall_GFactor.nii.gz: 40%|▍| 1.46M/3.64M [00:00<00:00, 4.
sub-MPI2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/15.5M [00:00<00:26, 615kB/s
sub-MPI2_acq-famp-0.66_TB1DREAM.nii.gz: 88%|▉| 77.0k/88.0k [00:00<00:00, 595kB/
sub-MPI2_acq-coilQaSagSmall_GFactor.nii.gz: 98%|▉| 3.56M/3.64M [00:00<00:00, 9.
sub-MPI2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/88.3k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/15.5M [00:00<00:11, 1.42MB/s
sub-MPI2_acq-famp_TB1DREAM.nii.gz: 95%|███▊| 83.6k/88.3k [00:00<00:00, 762kB/s]
sub-MPI2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 9%| | 1.46M/15.5M [00:00<00:03, 4.50MB/
sub-MPI2_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 31%|▎| 4.74M/15.5M [00:00<00:00, 13.9MB/
sub-MPI2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/937k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 57%|▌| 8.80M/15.5M [00:00<00:00, 22.2MB/
sub-MPI2_acq-famp_TB1TFL.nii.gz: 9%|▋ | 85.2k/937k [00:00<00:01, 655kB/s]
sub-MPI2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/84.2k [00:00<?, ?B/s]
sub-MPI2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI2_acq-coilQaTra_GFactor.nii.gz: 82%|▊| 12.7M/15.5M [00:00<00:00, 26.8MB/
sub-MPI2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/88.4k [00:00<?, ?B/s]
sub-MPI2_acq-famp_TB1TFL.nii.gz: 44%|███ | 408k/937k [00:00<00:00, 1.59MB/s]
sub-MPI2_acq-refv-0.66_TB1DREAM.nii.gz: 99%|▉| 83.6k/84.2k [00:00<00:00, 614kB/
sub-MPI2_acq-refv_TB1DREAM.nii.gz: 87%|███▍| 77.0k/88.4k [00:00<00:00, 547kB/s]
sub-MPI3_T2starw.json: 0%| | 0.00/2.38k [00:00<?, ?B/s]
sub-MPI3_UNIT1.json: 0%| | 0.00/2.04k [00:00<?, ?B/s]
sub-MPI3_T2starw.nii.gz: 0%| | 0.00/1.30M [00:00<?, ?B/s]
sub-MPI3_inv-1_MP2RAGE.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI3_T2starw.nii.gz: 6%|▉ | 85.2k/1.30M [00:00<00:01, 664kB/s]
sub-MPI3_T2starw.nii.gz: 19%|██▋ | 256k/1.30M [00:00<00:00, 1.22MB/s]
sub-MPI3_UNIT1.nii.gz: 0%| | 0.00/25.5M [00:00<?, ?B/s]
sub-MPI3_inv-2_MP2RAGE.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.4M [00:00<?, ?B/s]
sub-MPI3_T2starw.nii.gz: 61%|████████▌ | 817k/1.30M [00:00<00:00, 3.14MB/s]
sub-MPI3_UNIT1.nii.gz: 0%| | 83.6k/25.5M [00:00<00:33, 792kB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/18.4M [00:00<00:34, 563kB/
sub-MPI3_UNIT1.nii.gz: 1%|▏ | 356k/25.5M [00:00<00:16, 1.57MB/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/20.1M [00:00<?, ?B/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 349k/18.4M [00:00<00:11, 1.58MB/
sub-MPI3_rec-uncombined10_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_UNIT1.nii.gz: 4%|▌ | 951k/25.5M [00:00<00:08, 3.21MB/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/20.1M [00:00<00:36, 567kB/
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 4%| | 774k/18.4M [00:00<00:06, 2.70MB/
sub-MPI3_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/999k [00:00<?, ?B/s]
sub-MPI3_UNIT1.nii.gz: 11%|█▋ | 2.92M/25.5M [00:00<00:02, 9.42MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 11%| | 2.07M/18.4M [00:00<00:02, 6.73MB
sub-MPI3_UNIT1.nii.gz: 26%|███▊ | 6.53M/25.5M [00:00<00:01, 18.9MB/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 349k/20.1M [00:00<00:14, 1.41MB/
sub-MPI3_rec-uncombined10_T2starw.nii.gz: 9%| | 94.0k/999k [00:00<00:01, 684kB
sub-MPI3_rec-uncombined11_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 23%|▏| 4.21M/18.4M [00:00<00:01, 12.1MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 4%| | 758k/20.1M [00:00<00:08, 2.44MB/
sub-MPI3_UNIT1.nii.gz: 33%|████▉ | 8.45M/25.5M [00:00<00:00, 18.6MB/s]
sub-MPI3_rec-uncombined10_T2starw.nii.gz: 38%|▍| 383k/999k [00:00<00:00, 1.52MB
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 30%|▎| 5.54M/18.4M [00:00<00:01, 12.7MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 9%| | 1.74M/20.1M [00:00<00:03, 5.22MB
sub-MPI3_UNIT1.nii.gz: 40%|██████ | 10.3M/25.5M [00:00<00:00, 18.1MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 38%|▍| 7.07M/18.4M [00:00<00:00, 13.7MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 12%| | 2.43M/20.1M [00:00<00:03, 5.91MB
sub-MPI3_UNIT1.nii.gz: 48%|███████▏ | 12.1M/25.5M [00:00<00:00, 16.8MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 46%|▍| 8.49M/18.4M [00:00<00:00, 14.1MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 19%|▏| 3.87M/20.1M [00:00<00:01, 8.84MB
sub-MPI3_UNIT1.nii.gz: 55%|████████▎ | 14.0M/25.5M [00:01<00:00, 17.7MB/s]
sub-MPI3_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/984k [00:00<?, ?B/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 56%|▌| 10.3M/18.4M [00:00<00:00, 15.5MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 29%|▎| 5.75M/20.1M [00:00<00:01, 12.2MB
sub-MPI3_UNIT1.nii.gz: 63%|█████████▍ | 16.0M/25.5M [00:01<00:00, 18.4MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 66%|▋| 12.2M/18.4M [00:01<00:00, 16.8MB
sub-MPI3_rec-uncombined11_T2starw.nii.gz: 10%| | 94.0k/984k [00:00<00:01, 689kB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 38%|▍| 7.59M/20.1M [00:00<00:00, 14.4MB
sub-MPI3_rec-uncombined12_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_UNIT1.nii.gz: 70%|██████████▍ | 17.7M/25.5M [00:01<00:00, 17.3MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 75%|▋| 13.8M/18.4M [00:01<00:00, 15.9MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 45%|▍| 9.00M/20.1M [00:00<00:00, 14.3MB
sub-MPI3_rec-uncombined11_T2starw.nii.gz: 41%|▍| 400k/984k [00:00<00:00, 1.60MB
sub-MPI3_UNIT1.nii.gz: 77%|███████████▍ | 19.5M/25.5M [00:01<00:00, 17.5MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 84%|▊| 15.5M/18.4M [00:01<00:00, 16.5MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 53%|▌| 10.7M/20.1M [00:01<00:00, 15.3MB
sub-MPI3_UNIT1.nii.gz: 83%|████████████▍ | 21.2M/25.5M [00:01<00:00, 16.5MB/s]
sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz: 93%|▉| 17.1M/18.4M [00:01<00:00, 16.1MB
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 61%|▌| 12.2M/20.1M [00:01<00:00, 15.2MB
sub-MPI3_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/994k [00:00<?, ?B/s]
sub-MPI3_UNIT1.nii.gz: 90%|█████████████▍ | 22.8M/25.5M [00:01<00:00, 16.3MB/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 68%|▋| 13.7M/20.1M [00:01<00:00, 15.5MB
sub-MPI3_UNIT1.nii.gz: 100%|██████████████▉| 25.4M/25.5M [00:01<00:00, 19.3MB/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 81%|▊| 16.2M/20.1M [00:01<00:00, 17.4MB
sub-MPI3_rec-uncombined12_T2starw.nii.gz: 8%| | 83.6k/994k [00:00<00:01, 608kB
sub-MPI3_rec-uncombined13_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_inv-2_part-mag_MP2RAGE.nii.gz: 99%|▉| 19.8M/20.1M [00:01<00:00, 23.2MB
sub-MPI3_rec-uncombined12_T2starw.nii.gz: 36%|▎| 356k/994k [00:00<00:00, 1.49MB
sub-MPI3_rec-uncombined14_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/968k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined15_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined13_T2starw.nii.gz: 10%| | 94.0k/968k [00:00<00:01, 725kB
sub-MPI3_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/986k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined13_T2starw.nii.gz: 41%|▍| 400k/968k [00:00<00:00, 1.70MB
sub-MPI3_rec-uncombined14_T2starw.nii.gz: 8%| | 83.5k/1.03M [00:00<00:01, 613k
sub-MPI3_rec-uncombined15_T2starw.nii.gz: 8%| | 83.5k/986k [00:00<00:01, 615kB
sub-MPI3_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/1.02M [00:00<00:01, 652k
sub-MPI3_rec-uncombined16_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined14_T2starw.nii.gz: 32%|▎| 340k/1.03M [00:00<00:00, 1.63M
sub-MPI3_rec-uncombined15_T2starw.nii.gz: 39%|▍| 390k/986k [00:00<00:00, 1.89MB
sub-MPI3_rec-uncombined14_T2starw.nii.gz: 78%|▊| 832k/1.03M [00:00<00:00, 2.94M
sub-MPI3_rec-uncombined16_T2starw.nii.gz: 34%|▎| 356k/1.02M [00:00<00:00, 1.52M
sub-MPI3_rec-uncombined15_T2starw.nii.gz: 84%|▊| 832k/986k [00:00<00:00, 2.75MB
sub-MPI3_rec-uncombined17_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined18_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 606k
sub-MPI3_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined17_T2starw.nii.gz: 34%|▎| 356k/1.04M [00:00<00:00, 1.51M
sub-MPI3_rec-uncombined19_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined18_T2starw.nii.gz: 7%| | 75.3k/1.07M [00:00<00:01, 590k
sub-MPI3_rec-uncombined19_T2starw.nii.gz: 8%| | 83.5k/1.08M [00:00<00:01, 573k
sub-MPI3_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined18_T2starw.nii.gz: 32%|▎| 347k/1.07M [00:00<00:00, 1.50M
sub-MPI3_rec-uncombined19_T2starw.nii.gz: 32%|▎| 356k/1.08M [00:00<00:00, 1.43M
sub-MPI3_rec-uncombined1_T2starw.nii.gz: 8%| | 83.6k/1.02M [00:00<00:01, 649kB
sub-MPI3_rec-uncombined1_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined20_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined1_T2starw.nii.gz: 26%|▎| 272k/1.02M [00:00<00:00, 1.27MB
sub-MPI3_rec-uncombined1_T2starw.nii.gz: 97%|▉| 0.99M/1.02M [00:00<00:00, 3.93M
sub-MPI3_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined21_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined22_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/1.07M [00:00<00:01, 602k
sub-MPI3_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined20_T2starw.nii.gz: 33%|▎| 356k/1.07M [00:00<00:00, 1.52M
sub-MPI3_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 611k
sub-MPI3_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined22_T2starw.nii.gz: 8%| | 83.6k/1.07M [00:00<00:01, 649k
sub-MPI3_rec-uncombined23_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined21_T2starw.nii.gz: 36%|▎| 390k/1.05M [00:00<00:00, 1.56M
sub-MPI3_rec-uncombined22_T2starw.nii.gz: 32%|▎| 356k/1.07M [00:00<00:00, 1.37M
sub-MPI3_rec-uncombined23_T2starw.nii.gz: 7%| | 77.0k/1.07M [00:00<00:02, 494k
sub-MPI3_rec-uncombined24_T2starw.json: 0%| | 0.00/2.40k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined23_T2starw.nii.gz: 32%|▎| 349k/1.07M [00:00<00:00, 1.36M
sub-MPI3_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined24_T2starw.nii.gz: 8%| | 85.2k/1.01M [00:00<00:01, 646k
sub-MPI3_rec-uncombined2_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined3_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined24_T2starw.nii.gz: 29%|▎| 306k/1.01M [00:00<00:00, 1.13M
sub-MPI3_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/992k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined4_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined2_T2starw.nii.gz: 8%| | 83.6k/992k [00:00<00:01, 562kB/
sub-MPI3_rec-uncombined3_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 611kB
sub-MPI3_rec-uncombined2_T2starw.nii.gz: 36%|▎| 356k/992k [00:00<00:00, 1.56MB/
sub-MPI3_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/984k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined5_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined3_T2starw.nii.gz: 33%|▎| 339k/1.00M [00:00<00:00, 1.27MB
sub-MPI3_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined4_T2starw.nii.gz: 8%| | 83.6k/984k [00:00<00:01, 620kB/
sub-MPI3_rec-uncombined5_T2starw.nii.gz: 7%| | 77.0k/1.01M [00:00<00:01, 566kB
sub-MPI3_rec-uncombined4_T2starw.nii.gz: 36%|▎| 356k/984k [00:00<00:00, 1.47MB/
sub-MPI3_rec-uncombined6_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined4_T2starw.nii.gz: 98%|▉| 968k/984k [00:00<00:00, 3.29MB/
sub-MPI3_rec-uncombined7_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/992k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined5_T2starw.nii.gz: 34%|▎| 349k/1.01M [00:00<00:00, 1.26MB
sub-MPI3_rec-uncombined6_T2starw.nii.gz: 8%| | 83.5k/992k [00:00<00:01, 600kB/
sub-MPI3_rec-uncombined8_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined6_T2starw.nii.gz: 36%|▎| 356k/992k [00:00<00:00, 1.40MB/
sub-MPI3_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined9_T2starw.json: 0%| | 0.00/2.39k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined6_T2starw.nii.gz: 92%|▉| 917k/992k [00:00<00:00, 3.02MB/
sub-MPI3_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined7_T2starw.nii.gz: 8%| | 83.6k/1.03M [00:00<00:01, 611kB
sub-MPI3_rec-uncombined8_T2starw.nii.gz: 8%| | 83.5k/0.99M [00:00<00:01, 614kB
sub-MPI3_rec-uncombined9_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-MPI3_rec-uncombined7_T2starw.nii.gz: 34%|▎| 356k/1.03M [00:00<00:00, 1.42MB
sub-MPI3_rec-uncombined8_T2starw.nii.gz: 40%|▍| 407k/0.99M [00:00<00:00, 1.64MB
sub-MPI3_acq-anat_TB1TFL.json: 0%| | 0.00/2.62k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined9_T2starw.nii.gz: 8%| | 85.2k/0.99M [00:00<00:01, 553kB
sub-MPI3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/843k [00:00<?, ?B/s]
sub-MPI3_rec-uncombined9_T2starw.nii.gz: 35%|▎| 357k/0.99M [00:00<00:00, 1.46MB
sub-MPI3_acq-anat_TB1TFL.nii.gz: 10%|▋ | 83.6k/843k [00:00<00:01, 610kB/s]
sub-MPI3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.96M [00:00<?, ?B/s]
sub-MPI3_acq-anat_TB1TFL.nii.gz: 42%|██▉ | 356k/843k [00:00<00:00, 1.37MB/s]
sub-MPI3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/3.96M [00:00<00:06, 614kB/
sub-MPI3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.58M [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 339k/3.96M [00:00<00:03, 1.26MB/
sub-MPI3_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.58M [00:00<00:05, 61
sub-MPI3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz: 30%|▎| 1.19M/3.96M [00:00<00:00, 3.98MB
sub-MPI3_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.58M [00:00<00:02, 1.4
sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz: 88%|▉| 3.49M/3.96M [00:00<00:00, 10.7MB
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.6M [00:00<?, ?B/s]
sub-MPI3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.0k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaSagSmall_GFactor.nii.gz: 41%|▍| 1.46M/3.58M [00:00<00:00, 4.
sub-MPI3_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 1%| | 85.2k/15.6M [00:00<00:25, 632kB/s
sub-MPI3_acq-famp-0.66_TB1DREAM.nii.gz: 96%|▉| 83.6k/87.0k [00:00<00:00, 573kB/
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 2%| | 306k/15.6M [00:00<00:11, 1.42MB/s
sub-MPI3_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/88.9k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.21M/15.6M [00:00<00:03, 4.79MB/
sub-MPI3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.05k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 13%|▏| 1.96M/15.6M [00:00<00:02, 5.92MB/
sub-MPI3_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-MPI3_acq-famp-1.5_TB1DREAM.nii.gz: 96%|▉| 85.2k/88.9k [00:00<00:00, 619kB/s
sub-MPI3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/88.3k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 28%|▎| 4.34M/15.6M [00:00<00:00, 12.5MB/
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 57%|▌| 8.80M/15.6M [00:00<00:00, 23.0MB/
sub-MPI3_acq-famp_TB1DREAM.nii.gz: 97%|███▊| 85.2k/88.3k [00:00<00:00, 625kB/s]
sub-MPI3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-MPI3_acq-coilQaTra_GFactor.nii.gz: 72%|▋| 11.3M/15.6M [00:00<00:00, 23.9MB/
sub-MPI3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/82.6k [00:00<?, ?B/s]
sub-MPI3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/955k [00:00<?, ?B/s]
sub-MPI3_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 82.6k/82.6k [00:00<00:00, 651kB/
sub-MPI3_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/955k [00:00<00:01, 598kB/s]
sub-MPI3_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-MPI3_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/91.1k [00:00<?, ?B/s]
sub-MPI3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.07k [00:00<?, ?B/s]
sub-MPI3_acq-famp_TB1TFL.nii.gz: 36%|██▍ | 340k/955k [00:00<00:00, 1.32MB/s]
sub-MPI3_acq-refv-1.5_TB1DREAM.nii.gz: 92%|▉| 83.5k/91.1k [00:00<00:00, 529kB/s
sub-MPI3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/88.6k [00:00<?, ?B/s]
sub-MSSM1_T2starw.json: 0%| | 0.00/1.85k [00:00<?, ?B/s]
sub-MPI3_acq-refv_TB1DREAM.nii.gz: 94%|███▊| 83.6k/88.6k [00:00<00:00, 561kB/s]
sub-MSSM1_UNIT1.json: 0%| | 0.00/1.14k [00:00<?, ?B/s]
sub-MSSM1_inv-1_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM1_T2starw.nii.gz: 0%| | 0.00/1.78M [00:00<?, ?B/s]
sub-MSSM1_inv-2_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM1_T2starw.nii.gz: 5%|▌ | 83.6k/1.78M [00:00<00:03, 583kB/s]
sub-MSSM1_UNIT1.nii.gz: 0%| | 0.00/26.6M [00:00<?, ?B/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.7M [00:00<?, ?B/s]
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/20.7M [00:00<?, ?B/s]
sub-MSSM1_T2starw.nii.gz: 20%|██▌ | 356k/1.78M [00:00<00:01, 1.44MB/s]
sub-MSSM1_UNIT1.nii.gz: 0%| | 83.6k/26.6M [00:00<00:42, 651kB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/18.7M [00:00<00:31, 627kB
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.5k/20.7M [00:00<00:35, 615kB
sub-MSSM1_T2starw.nii.gz: 82%|█████████▊ | 1.46M/1.78M [00:00<00:00, 4.56MB/s]
sub-MSSM1_rec-uncombined00001_T2starw.nii.gz: 0%| | 0.00/374k [00:00<?, ?B/s]
sub-MSSM1_UNIT1.nii.gz: 1%|▏ | 356k/26.6M [00:00<00:19, 1.39MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 357k/18.7M [00:00<00:13, 1.43MB
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 407k/20.7M [00:00<00:12, 1.65MB
sub-MSSM1_UNIT1.nii.gz: 5%|▊ | 1.46M/26.6M [00:00<00:05, 5.00MB/s]
sub-MSSM1_rec-uncombined00001_T2starw.nii.gz: 23%|▏| 85.2k/374k [00:00<00:00, 6
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 8%| | 1.45M/18.7M [00:00<00:03, 4.54M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.33M/20.7M [00:00<00:04, 4.69M
sub-MSSM1_UNIT1.nii.gz: 14%|█▉ | 3.61M/26.6M [00:00<00:02, 10.9MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 17%|▏| 3.19M/18.7M [00:00<00:01, 8.91M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 11%| | 2.22M/20.7M [00:00<00:03, 6.33M
sub-MSSM1_rec-uncombined00001_T2starw.nii.gz: 82%|▊| 306k/374k [00:00<00:00, 1.
sub-MSSM1_UNIT1.nii.gz: 19%|██▌ | 4.98M/26.6M [00:00<00:01, 11.8MB/s]
sub-MSSM1_rec-uncombined00002_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 22%|▏| 4.14M/18.7M [00:00<00:01, 9.08M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 3.05M/20.7M [00:00<00:02, 7.11M
sub-MSSM1_UNIT1.nii.gz: 24%|███▎ | 6.40M/26.6M [00:00<00:01, 12.8MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 33%|▎| 6.10M/18.7M [00:00<00:01, 12.5M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.99M/20.7M [00:00<00:01, 11.3M
sub-MSSM1_UNIT1.nii.gz: 31%|████▎ | 8.25M/26.6M [00:00<00:01, 14.8MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 42%|▍| 7.96M/18.7M [00:00<00:00, 14.6M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 33%|▎| 6.91M/20.7M [00:00<00:01, 14.1M
sub-MSSM1_UNIT1.nii.gz: 38%|█████▎ | 10.2M/26.6M [00:00<00:01, 16.5MB/s]
sub-MSSM1_rec-uncombined00001_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00003_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 50%|▌| 9.42M/18.7M [00:00<00:00, 13.5M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 40%|▍| 8.30M/20.7M [00:00<00:00, 13.1M
sub-MSSM1_UNIT1.nii.gz: 44%|██████▏ | 11.8M/26.6M [00:01<00:01, 14.9MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 61%|▌| 11.4M/18.7M [00:01<00:00, 15.4M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 49%|▍| 10.2M/20.7M [00:01<00:00, 15.0M
sub-MSSM1_UNIT1.nii.gz: 51%|███████▏ | 13.7M/26.6M [00:01<00:00, 16.1MB/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 71%|▋| 13.3M/18.7M [00:01<00:00, 16.8M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 58%|▌| 12.1M/20.7M [00:01<00:00, 16.4M
sub-MSSM1_UNIT1.nii.gz: 58%|████████▏ | 15.6M/26.6M [00:01<00:00, 17.2MB/s]
sub-MSSM1_rec-uncombined00002_T2starw.nii.gz: 0%| | 0.00/378k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00005_T2starw.nii.gz: 0%| | 0.00/759k [00:00<?, ?B/s]
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 81%|▊| 15.1M/18.7M [00:01<00:00, 17.3M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 68%|▋| 14.0M/20.7M [00:01<00:00, 17.4M
sub-MSSM1_UNIT1.nii.gz: 65%|█████████▏ | 17.4M/26.6M [00:01<00:00, 17.8MB/s]
sub-MSSM1_rec-uncombined00002_T2starw.nii.gz: 23%|▏| 85.2k/378k [00:00<00:00, 6
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 91%|▉| 17.0M/18.7M [00:01<00:00, 18.0M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 77%|▊| 15.8M/20.7M [00:01<00:00, 18.0M
sub-MSSM1_rec-uncombined00005_T2starw.nii.gz: 11%| | 83.5k/759k [00:00<00:01, 6
sub-MSSM1_UNIT1.nii.gz: 72%|██████████ | 19.2M/26.6M [00:01<00:00, 17.9MB/s]
sub-MSSM1_rec-uncombined00005_T2starw.nii.gz: 36%|▎| 271k/759k [00:00<00:00, 1.
sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz: 100%|▉| 18.7M/18.7M [00:01<00:00, 16.6M
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 85%|▊| 17.6M/20.7M [00:01<00:00, 15.5M
sub-MSSM1_UNIT1.nii.gz: 78%|██████████▉ | 20.9M/26.6M [00:01<00:00, 14.8MB/s]
sub-MSSM1_inv-2_part-mag_MP2RAGE.nii.gz: 94%|▉| 19.4M/20.7M [00:01<00:00, 16.5M
sub-MSSM1_UNIT1.nii.gz: 87%|████████████▏ | 23.1M/26.6M [00:01<00:00, 16.9MB/s]
sub-MSSM1_rec-uncombined00005_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00003_T2starw.nii.gz: 0%| | 0.00/756k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00007_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00003_T2starw.nii.gz: 11%| | 85.2k/756k [00:00<00:00, 7
sub-MSSM1_rec-uncombined00003_T2starw.nii.gz: 47%|▍| 357k/756k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00007_T2starw.nii.gz: 0%| | 0.00/768k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00011_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00009_T2starw.nii.gz: 0%| | 0.00/770k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00009_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00007_T2starw.nii.gz: 11%| | 83.6k/768k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00009_T2starw.nii.gz: 11%| | 83.6k/770k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00007_T2starw.nii.gz: 46%|▍| 356k/768k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00009_T2starw.nii.gz: 46%|▍| 356k/770k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00013_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00011_T2starw.nii.gz: 0%| | 0.00/760k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00014_T2starw.nii.gz: 0%| | 0.00/371k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00011_T2starw.nii.gz: 11%| | 83.6k/760k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00014_T2starw.nii.gz: 23%|▏| 85.2k/371k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00014_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00011_T2starw.nii.gz: 47%|▍| 356k/760k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00013_T2starw.nii.gz: 0%| | 0.00/367k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00014_T2starw.nii.gz: 92%|▉| 340k/371k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00015_T2starw.nii.gz: 0%| | 0.00/743k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00013_T2starw.nii.gz: 23%|▏| 83.6k/367k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00015_T2starw.nii.gz: 10%| | 77.0k/743k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00015_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00013_T2starw.nii.gz: 78%|▊| 288k/367k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00015_T2starw.nii.gz: 36%|▎| 264k/743k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00019_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00017_T2starw.nii.gz: 0%| | 0.00/746k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00017_T2starw.nii.gz: 11%| | 83.6k/746k [00:00<00:01, 5
sub-MSSM1_rec-uncombined00021_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00017_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00017_T2starw.nii.gz: 27%|▎| 203k/746k [00:00<00:00, 89
sub-MSSM1_rec-uncombined00019_T2starw.nii.gz: 0%| | 0.00/759k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00021_T2starw.nii.gz: 0%| | 0.00/759k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00017_T2starw.nii.gz: 100%|█| 746k/746k [00:00<00:00, 2.
sub-MSSM1_rec-uncombined00019_T2starw.nii.gz: 11%| | 83.6k/759k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00023_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00021_T2starw.nii.gz: 11%| | 85.2k/759k [00:00<00:01, 5
sub-MSSM1_rec-uncombined00019_T2starw.nii.gz: 47%|▍| 356k/759k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00021_T2starw.nii.gz: 49%|▍| 375k/759k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00025_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00023_T2starw.nii.gz: 0%| | 0.00/737k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00025_T2starw.nii.gz: 0%| | 0.00/356k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00023_T2starw.nii.gz: 12%| | 85.2k/737k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00027_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00025_T2starw.nii.gz: 23%|▏| 83.6k/356k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00026_T2starw.nii.gz: 0%| | 0.00/360k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00026_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00023_T2starw.nii.gz: 48%|▍| 357k/737k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00025_T2starw.nii.gz: 95%|▉| 340k/356k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00026_T2starw.nii.gz: 23%|▏| 83.6k/360k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00026_T2starw.nii.gz: 99%|▉| 356k/360k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00027_T2starw.nii.gz: 0%| | 0.00/722k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00029_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00031_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00027_T2starw.nii.gz: 12%| | 83.5k/722k [00:00<00:01, 5
sub-MSSM1_rec-uncombined00031_T2starw.nii.gz: 0%| | 0.00/736k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00027_T2starw.nii.gz: 49%|▍| 356k/722k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00033_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00029_T2starw.nii.gz: 0%| | 0.00/722k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00031_T2starw.nii.gz: 12%| | 85.2k/736k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00033_T2starw.nii.gz: 0%| | 0.00/736k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00029_T2starw.nii.gz: 12%| | 83.6k/722k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00031_T2starw.nii.gz: 49%|▍| 357k/736k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00035_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00029_T2starw.nii.gz: 38%|▍| 272k/722k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00033_T2starw.nii.gz: 13%|▏| 94.0k/736k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00035_T2starw.nii.gz: 0%| | 0.00/706k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00033_T2starw.nii.gz: 54%|▌| 400k/736k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00035_T2starw.nii.gz: 12%| | 83.6k/706k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00037_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00035_T2starw.nii.gz: 50%|▌| 356k/706k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00038_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00037_T2starw.nii.gz: 0%| | 0.00/346k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00038_T2starw.nii.gz: 0%| | 0.00/351k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00037_T2starw.nii.gz: 24%|▏| 83.6k/346k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00038_T2starw.nii.gz: 24%|▏| 85.2k/351k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00039_T2starw.nii.gz: 0%| | 0.00/701k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00039_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00041_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00039_T2starw.nii.gz: 9%| | 66.5k/701k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00039_T2starw.nii.gz: 24%|▏| 169k/701k [00:00<00:00, 85
sub-MSSM1_rec-uncombined00043_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00041_T2starw.nii.gz: 0%| | 0.00/703k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00045_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00043_T2starw.nii.gz: 0%| | 0.00/713k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00041_T2starw.nii.gz: 12%| | 83.6k/703k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00043_T2starw.nii.gz: 12%| | 83.6k/713k [00:00<00:01, 6
sub-MSSM1_rec-uncombined00041_T2starw.nii.gz: 51%|▌| 356k/703k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00047_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00045_T2starw.nii.gz: 0%| | 0.00/712k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00043_T2starw.nii.gz: 33%|▎| 237k/713k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00047_T2starw.nii.gz: 0%| | 0.00/678k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00045_T2starw.nii.gz: 12%| | 83.6k/712k [00:00<00:00, 7
sub-MSSM1_rec-uncombined00047_T2starw.nii.gz: 14%|▏| 92.3k/678k [00:00<00:00, 7
sub-MSSM1_rec-uncombined00045_T2starw.nii.gz: 50%|▍| 356k/712k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00049_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00049_T2starw.nii.gz: 0%| | 0.00/334k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00047_T2starw.nii.gz: 59%|▌| 399k/678k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00050_T2starw.json: 0%| | 0.00/1.28k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00049_T2starw.nii.gz: 13%|▏| 42.9k/334k [00:00<00:00, 3
sub-MSSM1_rec-uncombined00049_T2starw.nii.gz: 76%|▊| 255k/334k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00050_T2starw.nii.gz: 0%| | 0.00/339k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00051_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00053_T2starw.json: 0%| | 0.00/1.31k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00050_T2starw.nii.gz: 25%|▏| 83.6k/339k [00:00<00:00, 5
sub-MSSM1_rec-uncombined00051_T2starw.nii.gz: 0%| | 0.00/677k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00053_T2starw.nii.gz: 0%| | 0.00/680k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00051_T2starw.nii.gz: 12%| | 78.0k/677k [00:00<00:01, 5
sub-MSSM1_rec-uncombined00055_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00053_T2starw.nii.gz: 13%|▏| 85.2k/680k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00055_T2starw.nii.gz: 0%| | 0.00/687k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00051_T2starw.nii.gz: 52%|▌| 349k/677k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00057_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00053_T2starw.nii.gz: 33%|▎| 221k/680k [00:00<00:00, 96
sub-MSSM1_rec-uncombined00055_T2starw.nii.gz: 11%| | 77.0k/687k [00:00<00:01, 5
sub-MSSM1_rec-uncombined00055_T2starw.nii.gz: 53%|▌| 366k/687k [00:00<00:00, 1.
sub-MSSM1_rec-uncombined00057_T2starw.nii.gz: 0%| | 0.00/685k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00059_T2starw.json: 0%| | 0.00/1.32k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00059_T2starw.nii.gz: 0%| | 0.00/651k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00057_T2starw.nii.gz: 11%| | 77.0k/685k [00:00<00:01, 5
sub-MSSM1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00059_T2starw.nii.gz: 13%|▏| 83.6k/651k [00:00<00:00, 6
sub-MSSM1_rec-uncombined00057_T2starw.nii.gz: 53%|▌| 366k/685k [00:00<00:00, 1.
sub-MSSM1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM1_rec-uncombined00059_T2starw.nii.gz: 55%|▌| 356k/651k [00:00<00:00, 1.
sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.34M [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.34M [00:00<00:07, 615kB
sub-MSSM1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.61M [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.34M [00:00<00:02, 1.43MB
sub-MSSM1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/93.1k [00:00<?, ?B/s]
sub-MSSM1_acq-famp_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.1M [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 85.2k/3.61M [00:00<00:06, 5
sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz: 34%|▎| 1.46M/4.34M [00:00<00:00, 4.60M
sub-MSSM1_acq-famp_TB1DREAM.nii.gz: 92%|██▋| 85.2k/93.1k [00:00<00:00, 625kB/s]
sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz: 99%|▉| 4.31M/4.34M [00:00<00:00, 11.9M
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.5k/16.1M [00:00<00:29, 568kB/
sub-MSSM1_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 357k/3.61M [00:00<00:02, 1.
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 2%| | 407k/16.1M [00:00<00:09, 1.78MB/
sub-MSSM1_acq-coilQaSagSmall_GFactor.nii.gz: 40%|▍| 1.46M/3.61M [00:00<00:00, 5
sub-MSSM1_acq-famp_TB1TFL.json: 0%| | 0.00/1.76k [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.28M/16.1M [00:00<00:03, 4.36MB
sub-MSSM1_acq-refv_TB1DREAM.json: 0%| | 0.00/1.36k [00:00<?, ?B/s]
sub-MSSM1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/830k [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 20%|▏| 3.14M/16.1M [00:00<00:01, 9.55MB
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 47%|▍| 7.59M/16.1M [00:00<00:00, 21.7MB
sub-MSSM1_acq-famp_TB1TFL.nii.gz: 10%|▌ | 83.6k/830k [00:00<00:01, 656kB/s]
sub-MSSM2_T2starw.json: 0%| | 0.00/1.85k [00:00<?, ?B/s]
sub-MSSM1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/93.3k [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 69%|▋| 11.1M/16.1M [00:00<00:00, 26.4MB
sub-MSSM1_acq-famp_TB1TFL.nii.gz: 43%|██▌ | 356k/830k [00:00<00:00, 1.52MB/s]
sub-MSSM2_T2starw.nii.gz: 0%| | 0.00/1.78M [00:00<?, ?B/s]
sub-MSSM1_acq-coilQaTra_GFactor.nii.gz: 98%|▉| 15.7M/16.1M [00:00<00:00, 33.4MB
sub-MSSM1_acq-refv_TB1DREAM.nii.gz: 90%|██▋| 83.5k/93.3k [00:00<00:00, 624kB/s]
sub-MSSM2_T2starw.nii.gz: 3%|▎ | 50.6k/1.78M [00:00<00:03, 464kB/s]
sub-MSSM2_T2starw.nii.gz: 13%|█▋ | 238k/1.78M [00:00<00:01, 1.18MB/s]
sub-MSSM2_UNIT1.json: 0%| | 0.00/1.14k [00:00<?, ?B/s]
sub-MSSM2_UNIT1.nii.gz: 0%| | 0.00/26.6M [00:00<?, ?B/s]
sub-MSSM2_T2starw.nii.gz: 29%|███▊ | 528k/1.78M [00:00<00:00, 1.94MB/s]
sub-MSSM2_inv-1_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM2_T2starw.nii.gz: 92%|██████████▉ | 1.63M/1.78M [00:00<00:00, 5.61MB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.4M [00:00<?, ?B/s]
sub-MSSM2_UNIT1.nii.gz: 0%| | 83.6k/26.6M [00:00<00:42, 648kB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.5k/17.4M [00:00<00:27, 652kB
sub-MSSM2_rec-uncombined00001_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_UNIT1.nii.gz: 1%|▏ | 356k/26.6M [00:00<00:19, 1.44MB/s]
sub-MSSM2_inv-2_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 407k/17.4M [00:00<00:10, 1.74MB
sub-MSSM2_UNIT1.nii.gz: 4%|▌ | 986k/26.6M [00:00<00:08, 3.33MB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 9%| | 1.58M/17.4M [00:00<00:02, 5.86M
sub-MSSM2_UNIT1.nii.gz: 10%|█▍ | 2.64M/26.6M [00:00<00:03, 8.33MB/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.0M [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00002_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 17%|▏| 2.90M/17.4M [00:00<00:01, 8.72M
sub-MSSM2_UNIT1.nii.gz: 18%|██▌ | 4.83M/26.6M [00:00<00:01, 13.2MB/s]
sub-MSSM2_rec-uncombined00001_T2starw.nii.gz: 0%| | 0.00/361k [00:00<?, ?B/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 31%|▎| 5.38M/17.4M [00:00<00:00, 14.6M
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/19.0M [00:00<00:35, 565kB
sub-MSSM2_UNIT1.nii.gz: 29%|████ | 7.64M/26.6M [00:00<00:01, 18.4MB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 42%|▍| 7.34M/17.4M [00:00<00:00, 16.5M
sub-MSSM2_UNIT1.nii.gz: 41%|█████▋ | 10.8M/26.6M [00:00<00:00, 23.0MB/s]
sub-MSSM2_rec-uncombined00001_T2starw.nii.gz: 18%|▏| 66.6k/361k [00:00<00:00, 5
sub-MSSM2_rec-uncombined00003_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 366k/19.0M [00:00<00:14, 1.34MB
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 51%|▌| 8.98M/17.4M [00:00<00:00, 16.4M
sub-MSSM2_UNIT1.nii.gz: 49%|██████▊ | 13.1M/26.6M [00:00<00:00, 21.1MB/s]
sub-MSSM2_rec-uncombined00001_T2starw.nii.gz: 89%|▉| 322k/361k [00:00<00:00, 1.
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.07M/19.0M [00:00<00:05, 3.36M
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 61%|▌| 10.6M/17.4M [00:00<00:00, 16.3M
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 11%| | 2.13M/19.0M [00:00<00:03, 5.87M
sub-MSSM2_UNIT1.nii.gz: 57%|███████▉ | 15.2M/26.6M [00:01<00:00, 19.0MB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 72%|▋| 12.5M/17.4M [00:00<00:00, 17.4M
sub-MSSM2_rec-uncombined00003_T2starw.nii.gz: 0%| | 0.00/712k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 20%|▏| 3.77M/19.0M [00:00<00:01, 9.46M
sub-MSSM2_UNIT1.nii.gz: 64%|█████████ | 17.2M/26.6M [00:01<00:00, 19.5MB/s]
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 84%|▊| 14.7M/17.4M [00:01<00:00, 19.1M
sub-MSSM2_rec-uncombined00002_T2starw.nii.gz: 0%| | 0.00/360k [00:00<?, ?B/s]
sub-MSSM2_UNIT1.nii.gz: 73%|██████████▎ | 19.5M/26.6M [00:01<00:00, 21.0MB/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 25%|▎| 4.83M/19.0M [00:00<00:01, 9.82M
sub-MSSM2_rec-uncombined00003_T2starw.nii.gz: 12%| | 83.6k/712k [00:00<00:00, 6
sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz: 97%|▉| 16.9M/17.4M [00:01<00:00, 20.1M
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 32%|▎| 6.02M/19.0M [00:00<00:01, 10.6M
sub-MSSM2_rec-uncombined00002_T2starw.nii.gz: 21%|▏| 77.0k/360k [00:00<00:00, 5
sub-MSSM2_UNIT1.nii.gz: 81%|███████████▎ | 21.6M/26.6M [00:01<00:00, 20.1MB/s]
sub-MSSM2_rec-uncombined00003_T2starw.nii.gz: 48%|▍| 339k/712k [00:00<00:00, 1.
sub-MSSM2_UNIT1.nii.gz: 91%|████████████▋ | 24.1M/26.6M [00:01<00:00, 21.8MB/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 39%|▍| 7.33M/19.0M [00:00<00:01, 10.9M
sub-MSSM2_rec-uncombined00005_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 44%|▍| 8.41M/19.0M [00:01<00:01, 9.87M
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 53%|▌| 9.99M/19.0M [00:01<00:00, 11.6M
sub-MSSM2_rec-uncombined00007_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00005_T2starw.nii.gz: 0%| | 0.00/728k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 60%|▌| 11.3M/19.0M [00:01<00:00, 12.2M
sub-MSSM2_rec-uncombined00009_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00005_T2starw.nii.gz: 11%| | 83.6k/728k [00:00<00:00, 6
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 66%|▋| 12.5M/19.0M [00:01<00:00, 11.8M
sub-MSSM2_rec-uncombined00007_T2starw.nii.gz: 0%| | 0.00/736k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 72%|▋| 13.7M/19.0M [00:01<00:00, 11.7M
sub-MSSM2_rec-uncombined00005_T2starw.nii.gz: 49%|▍| 356k/728k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00007_T2starw.nii.gz: 9%| | 67.5k/736k [00:00<00:01, 4
sub-MSSM2_rec-uncombined00009_T2starw.nii.gz: 0%| | 0.00/741k [00:00<?, ?B/s]
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 79%|▊| 14.9M/19.0M [00:01<00:00, 12.1M
sub-MSSM2_rec-uncombined00011_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00007_T2starw.nii.gz: 46%|▍| 339k/736k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00009_T2starw.nii.gz: 11%| | 83.6k/741k [00:00<00:01, 6
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 85%|▊| 16.1M/19.0M [00:01<00:00, 11.7M
sub-MSSM2_rec-uncombined00009_T2starw.nii.gz: 53%|▌| 390k/741k [00:00<00:00, 1.
sub-MSSM2_inv-2_part-mag_MP2RAGE.nii.gz: 93%|▉| 17.6M/19.0M [00:01<00:00, 10.7M
sub-MSSM2_rec-uncombined00013_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00011_T2starw.nii.gz: 0%| | 0.00/737k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00013_T2starw.nii.gz: 0%| | 0.00/354k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00014_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00011_T2starw.nii.gz: 11%| | 83.6k/737k [00:00<00:01, 6
sub-MSSM2_rec-uncombined00014_T2starw.nii.gz: 0%| | 0.00/352k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00013_T2starw.nii.gz: 24%|▏| 83.6k/354k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00011_T2starw.nii.gz: 48%|▍| 356k/737k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00015_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00014_T2starw.nii.gz: 24%|▏| 83.6k/352k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00013_T2starw.nii.gz: 100%|█| 354k/354k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00015_T2starw.nii.gz: 0%| | 0.00/696k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00014_T2starw.nii.gz: 96%|▉| 339k/352k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00017_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00015_T2starw.nii.gz: 12%| | 83.6k/696k [00:00<00:01, 5
sub-MSSM2_rec-uncombined00019_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00017_T2starw.nii.gz: 0%| | 0.00/702k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00015_T2starw.nii.gz: 51%|▌| 356k/696k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00021_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00017_T2starw.nii.gz: 12%| | 83.6k/702k [00:00<00:01, 6
sub-MSSM2_rec-uncombined00021_T2starw.nii.gz: 0%| | 0.00/711k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00017_T2starw.nii.gz: 48%|▍| 339k/702k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00023_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00019_T2starw.nii.gz: 0%| | 0.00/707k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00021_T2starw.nii.gz: 9%| | 66.6k/711k [00:00<00:01, 4
sub-MSSM2_rec-uncombined00023_T2starw.nii.gz: 0%| | 0.00/697k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00019_T2starw.nii.gz: 12%| | 83.6k/707k [00:00<00:00, 7
sub-MSSM2_rec-uncombined00021_T2starw.nii.gz: 45%|▍| 322k/711k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00023_T2starw.nii.gz: 12%| | 83.6k/697k [00:00<00:01, 6
sub-MSSM2_rec-uncombined00026_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00019_T2starw.nii.gz: 55%|▌| 390k/707k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00023_T2starw.nii.gz: 41%|▍| 289k/697k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00025_T2starw.nii.gz: 0%| | 0.00/345k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00025_T2starw.nii.gz: 24%|▏| 83.6k/345k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00025_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00027_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00026_T2starw.nii.gz: 0%| | 0.00/342k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00029_T2starw.nii.gz: 0%| | 0.00/678k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00026_T2starw.nii.gz: 24%|▏| 83.6k/342k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00029_T2starw.nii.gz: 12%| | 83.6k/678k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00027_T2starw.nii.gz: 0%| | 0.00/680k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00031_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00029_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00029_T2starw.nii.gz: 52%|▌| 356k/678k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00027_T2starw.nii.gz: 12%| | 83.6k/680k [00:00<00:01, 5
sub-MSSM2_rec-uncombined00027_T2starw.nii.gz: 57%|▌| 390k/680k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00031_T2starw.nii.gz: 0%| | 0.00/676k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00033_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00033_T2starw.nii.gz: 0%| | 0.00/682k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00031_T2starw.nii.gz: 12%| | 83.6k/676k [00:00<00:00, 7
sub-MSSM2_rec-uncombined00035_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00033_T2starw.nii.gz: 12%| | 83.6k/682k [00:00<00:01, 6
sub-MSSM2_rec-uncombined00031_T2starw.nii.gz: 53%|▌| 356k/676k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00037_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00033_T2starw.nii.gz: 50%|▍| 339k/682k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00035_T2starw.nii.gz: 0%| | 0.00/662k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00035_T2starw.nii.gz: 13%|▏| 83.6k/662k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00037_T2starw.nii.gz: 0%| | 0.00/335k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00038_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00035_T2starw.nii.gz: 51%|▌| 339k/662k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00037_T2starw.nii.gz: 25%|▏| 83.6k/335k [00:00<00:00, 5
sub-MSSM2_rec-uncombined00039_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00038_T2starw.nii.gz: 0%| | 0.00/333k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00038_T2starw.nii.gz: 25%|▎| 83.6k/333k [00:00<00:00, 7
sub-MSSM2_rec-uncombined00039_T2starw.nii.gz: 0%| | 0.00/662k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00041_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00043_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00041_T2starw.nii.gz: 0%| | 0.00/656k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00039_T2starw.nii.gz: 13%|▏| 83.6k/662k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00041_T2starw.nii.gz: 13%|▏| 83.6k/656k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00039_T2starw.nii.gz: 54%|▌| 356k/662k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00045_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00041_T2starw.nii.gz: 31%|▎| 204k/656k [00:00<00:00, 91
sub-MSSM2_rec-uncombined00043_T2starw.nii.gz: 0%| | 0.00/648k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00045_T2starw.nii.gz: 0%| | 0.00/655k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00043_T2starw.nii.gz: 13%|▏| 83.6k/648k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00045_T2starw.nii.gz: 13%|▏| 83.6k/655k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00047_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00043_T2starw.nii.gz: 55%|▌| 356k/648k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00047_T2starw.nii.gz: 0%| | 0.00/633k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00049_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00045_T2starw.nii.gz: 52%|▌| 339k/655k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00047_T2starw.nii.gz: 13%|▏| 83.6k/633k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00051_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00047_T2starw.nii.gz: 56%|▌| 356k/633k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00050_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00049_T2starw.nii.gz: 0%| | 0.00/328k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00050_T2starw.nii.gz: 0%| | 0.00/324k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00049_T2starw.nii.gz: 26%|▎| 83.6k/328k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00050_T2starw.nii.gz: 26%|▎| 83.6k/324k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00053_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00051_T2starw.nii.gz: 0%| | 0.00/647k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00053_T2starw.nii.gz: 0%| | 0.00/639k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00051_T2starw.nii.gz: 13%|▏| 83.6k/647k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00053_T2starw.nii.gz: 13%|▏| 83.6k/639k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00051_T2starw.nii.gz: 55%|▌| 356k/647k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00055_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00053_T2starw.nii.gz: 53%|▌| 339k/639k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00057_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00055_T2starw.nii.gz: 0%| | 0.00/627k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00055_T2starw.nii.gz: 13%|▏| 83.6k/627k [00:00<00:00, 7
sub-MSSM2_rec-uncombined00059_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00055_T2starw.nii.gz: 57%|▌| 356k/627k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00057_T2starw.nii.gz: 0%| | 0.00/633k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00059_T2starw.nii.gz: 0%| | 0.00/612k [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00057_T2starw.nii.gz: 13%|▏| 85.2k/633k [00:00<00:00, 6
sub-MSSM2_rec-uncombined00059_T2starw.nii.gz: 14%|▏| 83.6k/612k [00:00<00:00, 6
sub-MSSM2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.01M [00:00<?, ?B/s]
sub-MSSM2_rec-uncombined00057_T2starw.nii.gz: 56%|▌| 357k/633k [00:00<00:00, 1.
sub-MSSM2_rec-uncombined00059_T2starw.nii.gz: 58%|▌| 356k/612k [00:00<00:00, 1.
sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.01M [00:00<00:06, 595kB
sub-MSSM2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.61M [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaTra_GFactor.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 356k/4.01M [00:00<00:02, 1.38MB
sub-MSSM2_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 85.2k/3.61M [00:00<00:05, 6
sub-MSSM2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz: 32%|▎| 1.28M/4.01M [00:00<00:00, 4.25M
sub-MSSM2_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 357k/3.61M [00:00<00:02, 1.
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.8M [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz: 83%|▊| 3.33M/4.01M [00:00<00:00, 9.96M
sub-MSSM2_acq-coilQaSagSmall_GFactor.nii.gz: 33%|▎| 1.18M/3.61M [00:00<00:00, 3
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/15.8M [00:00<00:26, 615kB/
sub-MSSM2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/91.9k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaSagSmall_GFactor.nii.gz: 95%|▉| 3.44M/3.61M [00:00<00:00, 1
sub-MSSM2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 1%| | 186k/15.8M [00:00<00:20, 802kB/s
sub-MSSM2_acq-famp-0.66_TB1DREAM.nii.gz: 93%|▉| 85.2k/91.9k [00:00<00:00, 621kB
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 5%| | 747k/15.8M [00:00<00:05, 2.76MB/
sub-MSSM2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/91.2k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 14%|▏| 2.19M/15.8M [00:00<00:01, 7.31MB
sub-MSSM2_acq-famp_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM2_acq-famp_TB1DREAM.nii.gz: 92%|██▋| 83.6k/91.2k [00:00<00:00, 613kB/s]
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 23%|▏| 3.66M/15.8M [00:00<00:01, 9.58MB
sub-MSSM2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/90.5k [00:00<?, ?B/s]
sub-MSSM2_acq-famp_TB1TFL.json: 0%| | 0.00/1.74k [00:00<?, ?B/s]
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 48%|▍| 7.65M/15.8M [00:00<00:00, 19.8MB
sub-MSSM2_acq-famp-1.5_TB1DREAM.nii.gz: 92%|▉| 83.6k/90.5k [00:00<00:00, 612kB/
sub-MSSM2_acq-coilQaTra_GFactor.nii.gz: 71%|▋| 11.1M/15.8M [00:00<00:00, 25.0MB
sub-MSSM2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/794k [00:00<?, ?B/s]
sub-MSSM2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/1.36k [00:00<?, ?B/s]
sub-MSSM2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/84.1k [00:00<?, ?B/s]
sub-MSSM2_acq-famp_TB1TFL.nii.gz: 11%|▋ | 83.5k/794k [00:00<00:01, 652kB/s]
sub-MSSM2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/1.36k [00:00<?, ?B/s]
sub-MSSM2_acq-refv-0.66_TB1DREAM.nii.gz: 99%|▉| 83.6k/84.1k [00:00<00:00, 606kB
sub-MSSM2_acq-famp_TB1TFL.nii.gz: 51%|███ | 407k/794k [00:00<00:00, 1.74MB/s]
sub-MSSM2_acq-refv_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/86.8k [00:00<?, ?B/s]
sub-MSSM3_T2starw.json: 0%| | 0.00/1.75k [00:00<?, ?B/s]
sub-MSSM2_acq-refv-1.5_TB1DREAM.nii.gz: 96%|▉| 83.6k/86.8k [00:00<00:00, 546kB/
sub-MSSM3_T2starw.nii.gz: 0%| | 0.00/1.51M [00:00<?, ?B/s]
sub-MSSM3_UNIT1.json: 0%| | 0.00/1.14k [00:00<?, ?B/s]
sub-MSSM2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/85.5k [00:00<?, ?B/s]
sub-MSSM3_T2starw.nii.gz: 5%|▋ | 83.6k/1.51M [00:00<00:02, 659kB/s]
sub-MSSM2_acq-refv_TB1DREAM.nii.gz: 100%|██▉| 85.2k/85.5k [00:00<00:00, 751kB/s]
sub-MSSM3_inv-1_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM3_T2starw.nii.gz: 23%|██▉ | 356k/1.51M [00:00<00:00, 1.37MB/s]
sub-MSSM3_inv-2_MP2RAGE.json: 0%| | 0.00/1.18k [00:00<?, ?B/s]
sub-MSSM3_T2starw.nii.gz: 94%|███████████▎| 1.43M/1.51M [00:00<00:00, 4.91MB/s]
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.9M [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00001_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/18.9M [00:00<00:32, 609kB
sub-MSSM3_UNIT1.nii.gz: 0%| | 0.00/26.6M [00:00<?, ?B/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/22.0M [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00001_T2starw.nii.gz: 0%| | 0.00/371k [00:00<?, ?B/s]
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 356k/18.9M [00:00<00:13, 1.41MB
sub-MSSM3_UNIT1.nii.gz: 0%| | 83.6k/26.6M [00:00<00:46, 594kB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/22.0M [00:00<00:37, 610kB
sub-MSSM3_rec-uncombined00001_T2starw.nii.gz: 23%|▏| 83.6k/371k [00:00<00:00, 6
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.39M/18.9M [00:00<00:03, 4.98M
sub-MSSM3_UNIT1.nii.gz: 1%|▏ | 339k/26.6M [00:00<00:20, 1.33MB/s]
sub-MSSM3_rec-uncombined00002_T2starw.nii.gz: 0%| | 0.00/377k [00:00<?, ?B/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 356k/22.0M [00:00<00:16, 1.41MB
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 2.95M/18.9M [00:00<00:01, 8.60M
sub-MSSM3_rec-uncombined00001_T2starw.nii.gz: 96%|▉| 356k/371k [00:00<00:00, 1.
sub-MSSM3_UNIT1.nii.gz: 4%|▌ | 0.99M/26.6M [00:00<00:08, 3.23MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 4%| | 951k/22.0M [00:00<00:07, 3.15MB
sub-MSSM3_rec-uncombined00002_T2starw.nii.gz: 22%|▏| 83.6k/377k [00:00<00:00, 6
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.51M/18.9M [00:00<00:01, 11.1M
sub-MSSM3_UNIT1.nii.gz: 9%|█▎ | 2.40M/26.6M [00:00<00:03, 7.05MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 9%| | 2.06M/22.0M [00:00<00:03, 6.17M
sub-MSSM3_rec-uncombined00002_T2starw.nii.gz: 81%|▊| 305k/377k [00:00<00:00, 1.
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 33%|▎| 6.25M/18.9M [00:00<00:00, 13.3M
sub-MSSM3_UNIT1.nii.gz: 16%|██▏ | 4.15M/26.6M [00:00<00:02, 10.7MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 3.39M/22.0M [00:00<00:02, 8.68M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 40%|▍| 7.67M/18.9M [00:00<00:00, 13.8M
sub-MSSM3_rec-uncombined00002_T2starw.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_UNIT1.nii.gz: 20%|██▊ | 5.26M/26.6M [00:00<00:02, 10.7MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 22%|▏| 4.85M/22.0M [00:00<00:01, 10.8M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 48%|▍| 9.12M/18.9M [00:00<00:00, 14.2M
sub-MSSM3_UNIT1.nii.gz: 27%|███▊ | 7.13M/26.6M [00:00<00:01, 13.4MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 30%|▎| 6.68M/22.0M [00:00<00:01, 13.4M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 58%|▌| 11.0M/18.9M [00:00<00:00, 15.8M
sub-MSSM3_rec-uncombined00003_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_UNIT1.nii.gz: 32%|████▌ | 8.57M/26.6M [00:00<00:01, 13.9MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 37%|▎| 8.04M/22.0M [00:00<00:01, 13.7M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 66%|▋| 12.5M/18.9M [00:01<00:00, 15.6M
sub-MSSM3_rec-uncombined00007_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_UNIT1.nii.gz: 38%|█████▎ | 9.99M/26.6M [00:01<00:01, 14.2MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 43%|▍| 9.51M/22.0M [00:00<00:00, 14.2M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 74%|▋| 14.0M/18.9M [00:01<00:00, 15.6M
sub-MSSM3_UNIT1.nii.gz: 45%|██████▎ | 12.0M/26.6M [00:01<00:00, 16.1MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 11.4M/22.0M [00:01<00:00, 16.0M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 84%|▊| 15.9M/18.9M [00:01<00:00, 16.9M
sub-MSSM3_UNIT1.nii.gz: 52%|███████▎ | 13.9M/26.6M [00:01<00:00, 17.4MB/s]
sub-MSSM3_rec-uncombined00003_T2starw.nii.gz: 0%| | 0.00/759k [00:00<?, ?B/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 61%|▌| 13.4M/22.0M [00:01<00:00, 17.3M
sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz: 94%|▉| 17.8M/18.9M [00:01<00:00, 17.9M
sub-MSSM3_UNIT1.nii.gz: 60%|████████▎ | 15.9M/26.6M [00:01<00:00, 18.3MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 69%|▋| 15.1M/22.0M [00:01<00:00, 16.8M
sub-MSSM3_rec-uncombined00005_T2starw.nii.gz: 0%| | 0.00/766k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00003_T2starw.nii.gz: 11%| | 83.6k/759k [00:00<00:01, 6
sub-MSSM3_UNIT1.nii.gz: 67%|█████████▍ | 17.9M/26.6M [00:01<00:00, 19.1MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 80%|▊| 17.7M/22.0M [00:01<00:00, 19.8M
sub-MSSM3_rec-uncombined00005_T2starw.nii.gz: 11%| | 85.2k/766k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00003_T2starw.nii.gz: 47%|▍| 356k/759k [00:00<00:00, 1.
sub-MSSM3_UNIT1.nii.gz: 77%|██████████▋ | 20.4M/26.6M [00:01<00:00, 21.2MB/s]
sub-MSSM3_inv-2_part-mag_MP2RAGE.nii.gz: 91%|▉| 20.0M/22.0M [00:01<00:00, 21.2M
sub-MSSM3_rec-uncombined00005_T2starw.nii.gz: 47%|▍| 357k/766k [00:00<00:00, 1.
sub-MSSM3_UNIT1.nii.gz: 84%|███████████▊ | 22.4M/26.6M [00:01<00:00, 20.2MB/s]
sub-MSSM3_rec-uncombined00005_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_UNIT1.nii.gz: 92%|████████████▊ | 24.4M/26.6M [00:01<00:00, 18.8MB/s]
sub-MSSM3_rec-uncombined00007_T2starw.nii.gz: 0%| | 0.00/769k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00011_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00009_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00007_T2starw.nii.gz: 10%| | 77.0k/769k [00:00<00:01, 5
sub-MSSM3_rec-uncombined00009_T2starw.nii.gz: 0%| | 0.00/768k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00011_T2starw.nii.gz: 0%| | 0.00/762k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00007_T2starw.nii.gz: 48%|▍| 366k/769k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00009_T2starw.nii.gz: 11%| | 83.6k/768k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00011_T2starw.nii.gz: 11%| | 83.6k/762k [00:00<00:01, 5
sub-MSSM3_rec-uncombined00013_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00009_T2starw.nii.gz: 26%|▎| 204k/768k [00:00<00:00, 91
sub-MSSM3_rec-uncombined00014_T2starw.nii.gz: 0%| | 0.00/371k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00011_T2starw.nii.gz: 44%|▍| 339k/762k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00009_T2starw.nii.gz: 95%|▉| 730k/768k [00:00<00:00, 2.
sub-MSSM3_rec-uncombined00014_T2starw.nii.gz: 21%|▏| 76.5k/371k [00:00<00:00, 4
sub-MSSM3_rec-uncombined00014_T2starw.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00013_T2starw.nii.gz: 0%| | 0.00/367k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00013_T2starw.nii.gz: 23%|▏| 85.2k/367k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00015_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00017_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00015_T2starw.nii.gz: 0%| | 0.00/748k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00013_T2starw.nii.gz: 97%|▉| 357k/367k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00017_T2starw.nii.gz: 0%| | 0.00/755k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00015_T2starw.nii.gz: 10%| | 77.0k/748k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00017_T2starw.nii.gz: 11%| | 85.2k/755k [00:00<00:00, 7
sub-MSSM3_rec-uncombined00019_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00015_T2starw.nii.gz: 49%|▍| 366k/748k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00017_T2starw.nii.gz: 50%|▍| 375k/755k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00021_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00019_T2starw.nii.gz: 0%| | 0.00/756k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00019_T2starw.nii.gz: 11%| | 83.6k/756k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00021_T2starw.nii.gz: 0%| | 0.00/755k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00025_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00019_T2starw.nii.gz: 45%|▍| 339k/756k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00023_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00021_T2starw.nii.gz: 11%| | 83.6k/755k [00:00<00:01, 5
sub-MSSM3_rec-uncombined00023_T2starw.nii.gz: 0%| | 0.00/740k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00021_T2starw.nii.gz: 47%|▍| 356k/755k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00023_T2starw.nii.gz: 10%| | 77.0k/740k [00:00<00:01, 5
sub-MSSM3_rec-uncombined00025_T2starw.nii.gz: 0%| | 0.00/358k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00026_T2starw.nii.gz: 0%| | 0.00/363k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00023_T2starw.nii.gz: 52%|▌| 383k/740k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00025_T2starw.nii.gz: 24%|▏| 85.2k/358k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00027_T2starw.nii.gz: 0%| | 0.00/728k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00026_T2starw.nii.gz: 23%|▏| 83.6k/363k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00026_T2starw.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00025_T2starw.nii.gz: 100%|▉| 357k/358k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00027_T2starw.nii.gz: 11%| | 83.6k/728k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00026_T2starw.nii.gz: 61%|▌| 221k/363k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00027_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00027_T2starw.nii.gz: 53%|▌| 390k/728k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00029_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00033_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00031_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00029_T2starw.nii.gz: 0%| | 0.00/731k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00031_T2starw.nii.gz: 0%| | 0.00/735k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00029_T2starw.nii.gz: 11%| | 83.6k/731k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00033_T2starw.nii.gz: 0%| | 0.00/730k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00037_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00035_T2starw.nii.gz: 0%| | 0.00/705k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00031_T2starw.nii.gz: 12%| | 85.2k/735k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00029_T2starw.nii.gz: 49%|▍| 356k/731k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00033_T2starw.nii.gz: 11%| | 83.5k/730k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00035_T2starw.nii.gz: 12%| | 85.2k/705k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00031_T2starw.nii.gz: 56%|▌| 408k/735k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00033_T2starw.nii.gz: 56%|▌| 407k/730k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00035_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00035_T2starw.nii.gz: 51%|▌| 357k/705k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00038_T2starw.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00037_T2starw.nii.gz: 0%| | 0.00/347k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00041_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00038_T2starw.nii.gz: 0%| | 0.00/353k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00037_T2starw.nii.gz: 25%|▏| 85.2k/347k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00039_T2starw.nii.gz: 0%| | 0.00/708k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00038_T2starw.nii.gz: 27%|▎| 94.0k/353k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00039_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00039_T2starw.nii.gz: 12%| | 83.6k/708k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00041_T2starw.nii.gz: 0%| | 0.00/710k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00039_T2starw.nii.gz: 45%|▍| 322k/708k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00041_T2starw.nii.gz: 12%| | 83.6k/710k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00043_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00045_T2starw.nii.gz: 0%| | 0.00/705k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00041_T2starw.nii.gz: 50%|▌| 356k/710k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00043_T2starw.nii.gz: 0%| | 0.00/712k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00045_T2starw.nii.gz: 12%| | 83.5k/705k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00043_T2starw.nii.gz: 12%| | 83.6k/712k [00:00<00:01, 6
sub-MSSM3_rec-uncombined00047_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00045_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00045_T2starw.nii.gz: 50%|▌| 356k/705k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00043_T2starw.nii.gz: 52%|▌| 373k/712k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00047_T2starw.nii.gz: 0%| | 0.00/672k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00049_T2starw.json: 0%| | 0.00/1.21k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00047_T2starw.nii.gz: 10%| | 66.6k/672k [00:00<00:01, 4
sub-MSSM3_rec-uncombined00050_T2starw.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00049_T2starw.nii.gz: 0%| | 0.00/335k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00050_T2starw.nii.gz: 0%| | 0.00/342k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00047_T2starw.nii.gz: 48%|▍| 322k/672k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00049_T2starw.nii.gz: 23%|▏| 77.0k/335k [00:00<00:00, 5
sub-MSSM3_rec-uncombined00050_T2starw.nii.gz: 25%|▏| 85.2k/342k [00:00<00:00, 7
sub-MSSM3_rec-uncombined00051_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00051_T2starw.nii.gz: 0%| | 0.00/686k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00053_T2starw.nii.gz: 0%| | 0.00/686k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00051_T2starw.nii.gz: 12%| | 83.6k/686k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00053_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00055_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00053_T2starw.nii.gz: 7%| | 50.6k/686k [00:00<00:01, 4
sub-MSSM3_rec-uncombined00051_T2starw.nii.gz: 52%|▌| 356k/686k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00055_T2starw.nii.gz: 0%| | 0.00/687k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00053_T2starw.nii.gz: 25%|▏| 169k/686k [00:00<00:00, 88
sub-MSSM3_rec-uncombined00057_T2starw.json: 0%| | 0.00/1.26k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00055_T2starw.nii.gz: 11%| | 77.0k/687k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00059_T2starw.json: 0%| | 0.00/1.25k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00055_T2starw.nii.gz: 53%|▌| 366k/687k [00:00<00:00, 1.
sub-MSSM3_rec-uncombined00057_T2starw.nii.gz: 0%| | 0.00/676k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00059_T2starw.nii.gz: 0%| | 0.00/642k [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00057_T2starw.nii.gz: 12%| | 83.6k/676k [00:00<00:00, 6
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.32M [00:00<?, ?B/s]
sub-MSSM3_rec-uncombined00059_T2starw.nii.gz: 13%|▏| 83.6k/642k [00:00<00:00, 6
sub-MSSM3_rec-uncombined00057_T2starw.nii.gz: 53%|▌| 356k/676k [00:00<00:00, 1.
sub-MSSM3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.61M [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/1.29k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 77.0k/4.32M [00:00<00:08, 545kB
sub-MSSM3_rec-uncombined00059_T2starw.nii.gz: 55%|▌| 356k/642k [00:00<00:00, 1.
sub-MSSM3_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 78.0k/3.61M [00:00<00:05, 6
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 349k/4.32M [00:00<00:02, 1.40MB
sub-MSSM3_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 366k/3.61M [00:00<00:02, 1.
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 33%|▎| 1.44M/4.32M [00:00<00:00, 5.18M
sub-MSSM3_acq-coilQaTra_GFactor.json: 0%| | 0.00/1.22k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaSagSmall_GFactor.nii.gz: 31%|▎| 1.11M/3.61M [00:00<00:00, 3
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 47%|▍| 2.02M/4.32M [00:00<00:00, 5.49M
sub-MSSM3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.7k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/16.0M [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaSagSmall_GFactor.nii.gz: 76%|▊| 2.72M/3.61M [00:00<00:00, 8
sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz: 98%|▉| 4.25M/4.32M [00:00<00:00, 11.4M
sub-MSSM3_acq-famp-0.66_TB1DREAM.nii.gz: 92%|▉| 85.2k/92.7k [00:00<00:00, 572kB
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/16.0M [00:00<00:29, 570kB/
sub-MSSM3_acq-famp_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 2%| | 356k/16.0M [00:00<00:11, 1.42MB/
sub-MSSM3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 8%| | 1.33M/16.0M [00:00<00:03, 4.73MB
sub-MSSM3_acq-famp_TB1TFL.json: 0%| | 0.00/1.73k [00:00<?, ?B/s]
sub-MSSM3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.9k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 18%|▏| 2.93M/16.0M [00:00<00:01, 8.62MB
sub-MSSM3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/89.7k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 45%|▍| 7.23M/16.0M [00:00<00:00, 20.4MB
sub-MSSM3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/1.36k [00:00<?, ?B/s]
sub-MSSM3_acq-famp_TB1DREAM.nii.gz: 90%|██▋| 83.6k/92.9k [00:00<00:00, 604kB/s]
sub-MSSM3_acq-refv-0.66_TB1DREAM.nii.gz: 55%|▌| 49.6k/89.7k [00:00<00:00, 458kB
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 61%|▌| 9.85M/16.0M [00:00<00:00, 22.7MB
sub-MSSM3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/833k [00:00<?, ?B/s]
sub-MSSM3_acq-coilQaTra_GFactor.nii.gz: 84%|▊| 13.5M/16.0M [00:00<00:00, 26.4MB
sub-MSSM3_acq-famp_TB1TFL.nii.gz: 10%|▌ | 83.6k/833k [00:00<00:01, 611kB/s]
sub-MSSM3_acq-refv_TB1DREAM.json: 0%| | 0.00/1.35k [00:00<?, ?B/s]
sub-NTNU1_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-MSSM3_acq-famp_TB1TFL.nii.gz: 37%|██▏ | 306k/833k [00:00<00:00, 1.45MB/s]
sub-MSSM3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/92.7k [00:00<?, ?B/s]
sub-MSSM3_acq-famp_TB1TFL.nii.gz: 88%|█████▎| 730k/833k [00:00<00:00, 2.55MB/s]
sub-MSSM3_acq-refv_TB1DREAM.nii.gz: 90%|██▋| 83.6k/92.7k [00:00<00:00, 624kB/s]
sub-NTNU1_UNIT1.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU1_T2starw.nii.gz: 0%| | 0.00/1.33M [00:00<?, ?B/s]
sub-NTNU1_T2starw.nii.gz: 6%|▊ | 83.6k/1.33M [00:00<00:01, 658kB/s]
sub-NTNU1_UNIT1.nii.gz: 0%| | 0.00/26.0M [00:00<?, ?B/s]
sub-NTNU1_inv-1_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU1_T2starw.nii.gz: 26%|███▍ | 356k/1.33M [00:00<00:00, 1.46MB/s]
sub-NTNU1_inv-2_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.7M [00:00<?, ?B/s]
sub-NTNU1_UNIT1.nii.gz: 0%| | 83.6k/26.0M [00:00<00:50, 543kB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/19.7M [00:00<00:29, 704kB
sub-NTNU1_UNIT1.nii.gz: 1%|▏ | 356k/26.0M [00:00<00:18, 1.45MB/s]
sub-NTNU1_UNIT1.nii.gz: 5%|▋ | 1.21M/26.0M [00:00<00:05, 4.34MB/s]
sub-NTNU1_rec-uncombined01_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 356k/19.7M [00:00<00:14, 1.45MB
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/21.4M [00:00<?, ?B/s]
sub-NTNU1_UNIT1.nii.gz: 8%|█ | 2.08M/26.0M [00:00<00:04, 6.00MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.29M/19.7M [00:00<00:04, 4.65M
sub-NTNU1_UNIT1.nii.gz: 22%|███ | 5.64M/26.0M [00:00<00:01, 16.5MB/s]
sub-NTNU1_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/21.4M [00:00<00:35, 623kB
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 15%|▏| 2.87M/19.7M [00:00<00:02, 8.69M
sub-NTNU1_UNIT1.nii.gz: 32%|████▌ | 8.39M/26.0M [00:00<00:00, 20.4MB/s]
sub-NTNU1_rec-uncombined02_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 25%|▎| 4.96M/19.7M [00:00<00:01, 12.7M
sub-NTNU1_rec-uncombined01_T2starw.nii.gz: 8%| | 85.2k/1.06M [00:00<00:01, 565
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 408k/21.4M [00:00<00:13, 1.64MB
sub-NTNU1_UNIT1.nii.gz: 40%|█████▌ | 10.4M/26.0M [00:00<00:00, 18.9MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 35%|▎| 6.94M/19.7M [00:00<00:00, 15.2M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 5%| | 1.15M/21.4M [00:00<00:05, 3.95M
sub-NTNU1_rec-uncombined01_T2starw.nii.gz: 33%|▎| 357k/1.06M [00:00<00:00, 1.46
sub-NTNU1_UNIT1.nii.gz: 47%|██████▋ | 12.3M/26.0M [00:00<00:00, 18.9MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 44%|▍| 8.62M/19.7M [00:00<00:00, 15.9M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 10%| | 2.15M/21.4M [00:00<00:03, 6.22M
sub-NTNU1_rec-uncombined01_T2starw.nii.gz: 92%|▉| 0.98M/1.06M [00:00<00:00, 3.4
sub-NTNU1_UNIT1.nii.gz: 55%|███████▋ | 14.2M/26.0M [00:01<00:00, 17.6MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 52%|▌| 10.2M/19.7M [00:00<00:00, 15.5M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 17%|▏| 3.57M/21.4M [00:00<00:02, 9.09M
sub-NTNU1_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU1_UNIT1.nii.gz: 62%|████████▋ | 16.2M/26.0M [00:01<00:00, 18.5MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 62%|▌| 12.1M/19.7M [00:00<00:00, 17.0M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 25%|▎| 5.45M/21.4M [00:00<00:01, 12.5M
sub-NTNU1_rec-uncombined02_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 615
sub-NTNU1_UNIT1.nii.gz: 69%|█████████▋ | 18.0M/26.0M [00:01<00:00, 18.6MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 71%|▋| 14.0M/19.7M [00:01<00:00, 17.7M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 35%|▎| 7.40M/21.4M [00:00<00:00, 14.9M
sub-NTNU1_rec-uncombined03_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined02_T2starw.nii.gz: 35%|▎| 356k/1.01M [00:00<00:00, 1.42
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 42%|▍| 8.88M/21.4M [00:00<00:00, 14.6M
sub-NTNU1_UNIT1.nii.gz: 76%|██████████▋ | 19.8M/26.0M [00:01<00:00, 17.4MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 80%|▊| 15.7M/19.7M [00:01<00:00, 16.5M
sub-NTNU1_rec-uncombined02_T2starw.nii.gz: 97%|▉| 0.98M/1.01M [00:00<00:00, 3.3
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 49%|▍| 10.4M/21.4M [00:01<00:00, 14.2M
sub-NTNU1_UNIT1.nii.gz: 83%|███████████▌ | 21.5M/26.0M [00:01<00:00, 16.3MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 88%|▉| 17.3M/19.7M [00:01<00:00, 15.6M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 58%|▌| 12.4M/21.4M [00:01<00:00, 16.1M
sub-NTNU1_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-NTNU1_UNIT1.nii.gz: 90%|████████████▌ | 23.4M/26.0M [00:01<00:00, 17.1MB/s]
sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz: 98%|▉| 19.2M/19.7M [00:01<00:00, 16.8M
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 66%|▋| 14.2M/21.4M [00:01<00:00, 16.7M
sub-NTNU1_UNIT1.nii.gz: 97%|█████████████▌| 25.2M/26.0M [00:01<00:00, 17.8MB/s]
sub-NTNU1_rec-uncombined03_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 534
sub-NTNU1_rec-uncombined04_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 74%|▋| 15.8M/21.4M [00:01<00:00, 16.7M
sub-NTNU1_rec-uncombined03_T2starw.nii.gz: 33%|▎| 356k/1.04M [00:00<00:00, 1.47
sub-NTNU1_inv-2_part-mag_MP2RAGE.nii.gz: 94%|▉| 20.2M/21.4M [00:01<00:00, 25.3M
sub-NTNU1_rec-uncombined05_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined03_T2starw.nii.gz: 73%|▋| 781k/1.04M [00:00<00:00, 2.58
sub-NTNU1_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined06_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined05_T2starw.nii.gz: 8%| | 83.5k/1.03M [00:00<00:01, 520
sub-NTNU1_rec-uncombined04_T2starw.nii.gz: 8%| | 83.5k/1.02M [00:00<00:01, 612
sub-NTNU1_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined08_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined05_T2starw.nii.gz: 39%|▍| 407k/1.03M [00:00<00:00, 1.62
sub-NTNU1_rec-uncombined04_T2starw.nii.gz: 39%|▍| 407k/1.02M [00:00<00:00, 1.64
sub-NTNU1_rec-uncombined06_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 572
sub-NTNU1_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/1.09M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined06_T2starw.nii.gz: 33%|▎| 356k/1.04M [00:00<00:00, 1.43
sub-NTNU1_rec-uncombined07_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined07_T2starw.nii.gz: 7%| | 83.6k/1.09M [00:00<00:01, 610
sub-NTNU1_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined09_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined07_T2starw.nii.gz: 35%|▎| 390k/1.09M [00:00<00:00, 1.56
sub-NTNU1_rec-uncombined08_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 617
sub-NTNU1_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/1.09M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined08_T2starw.nii.gz: 34%|▎| 356k/1.01M [00:00<00:00, 1.43
sub-NTNU1_rec-uncombined10_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined09_T2starw.nii.gz: 8%| | 83.6k/1.08M [00:00<00:01, 706
sub-NTNU1_rec-uncombined10_T2starw.nii.gz: 8%| | 83.6k/1.09M [00:00<00:01, 589
sub-NTNU1_rec-uncombined09_T2starw.nii.gz: 32%|▎| 356k/1.08M [00:00<00:00, 1.49
sub-NTNU1_rec-uncombined11_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined10_T2starw.nii.gz: 32%|▎| 356k/1.09M [00:00<00:00, 1.39
sub-NTNU1_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/1.12M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/1.11M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined11_T2starw.nii.gz: 7%| | 77.0k/1.12M [00:00<00:01, 567
sub-NTNU1_rec-uncombined13_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined12_T2starw.nii.gz: 7%| | 83.6k/1.11M [00:00<00:01, 615
sub-NTNU1_rec-uncombined12_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined11_T2starw.nii.gz: 32%|▎| 366k/1.12M [00:00<00:00, 1.47
sub-NTNU1_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/1.09M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined12_T2starw.nii.gz: 31%|▎| 356k/1.11M [00:00<00:00, 1.40
sub-NTNU1_rec-uncombined13_T2starw.nii.gz: 7%| | 83.6k/1.09M [00:00<00:01, 611
sub-NTNU1_rec-uncombined14_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined13_T2starw.nii.gz: 30%|▎| 339k/1.09M [00:00<00:00, 1.34
sub-NTNU1_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.14M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined15_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined14_T2starw.nii.gz: 7%| | 85.2k/1.14M [00:00<00:01, 621
sub-NTNU1_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/1.10M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined14_T2starw.nii.gz: 35%|▎| 408k/1.14M [00:00<00:00, 1.64
sub-NTNU1_rec-uncombined17_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined15_T2starw.nii.gz: 7%| | 83.6k/1.10M [00:00<00:01, 544
sub-NTNU1_rec-uncombined16_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.09M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined15_T2starw.nii.gz: 30%|▎| 339k/1.10M [00:00<00:00, 1.38
sub-NTNU1_rec-uncombined16_T2starw.nii.gz: 7%| | 83.6k/1.09M [00:00<00:01, 608
sub-NTNU1_rec-uncombined18_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined16_T2starw.nii.gz: 32%|▎| 356k/1.09M [00:00<00:00, 1.41
sub-NTNU1_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined19_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined17_T2starw.nii.gz: 8%| | 85.2k/0.99M [00:00<00:01, 617
sub-NTNU1_rec-uncombined18_T2starw.nii.gz: 8%| | 77.0k/0.98M [00:00<00:01, 565
sub-NTNU1_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined17_T2starw.nii.gz: 33%|▎| 340k/0.99M [00:00<00:00, 1.36
sub-NTNU1_rec-uncombined18_T2starw.nii.gz: 36%|▎| 366k/0.98M [00:00<00:00, 1.47
sub-NTNU1_rec-uncombined20_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined19_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 539
sub-NTNU1_rec-uncombined18_T2starw.nii.gz: 95%|▉| 961k/0.98M [00:00<00:00, 3.21
sub-NTNU1_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined19_T2starw.nii.gz: 34%|▎| 356k/1.01M [00:00<00:00, 1.44
sub-NTNU1_rec-uncombined20_T2starw.nii.gz: 8%| | 85.2k/0.99M [00:00<00:01, 595
sub-NTNU1_rec-uncombined21_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined22_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined20_T2starw.nii.gz: 35%|▎| 357k/0.99M [00:00<00:00, 1.43
sub-NTNU1_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 667
sub-NTNU1_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined23_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined21_T2starw.nii.gz: 38%|▍| 390k/1.00M [00:00<00:00, 1.56
sub-NTNU1_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined22_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 533
sub-NTNU1_rec-uncombined23_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 572
sub-NTNU1_rec-uncombined24_T2starw.json: 0%| | 0.00/2.46k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined22_T2starw.nii.gz: 35%|▎| 356k/1.01M [00:00<00:00, 1.46
sub-NTNU1_rec-uncombined23_T2starw.nii.gz: 35%|▎| 356k/0.98M [00:00<00:00, 1.49
sub-NTNU1_acq-anat_TB1TFL.json: 0%| | 0.00/2.68k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined23_T2starw.nii.gz: 98%|▉| 986k/0.98M [00:00<00:00, 3.35
sub-NTNU1_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined24_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 613
sub-NTNU1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/846k [00:00<?, ?B/s]
sub-NTNU1_rec-uncombined24_T2starw.nii.gz: 33%|▎| 340k/1.01M [00:00<00:00, 1.35
sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.11M [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.23k [00:00<?, ?B/s]
sub-NTNU1_acq-anat_TB1TFL.nii.gz: 10%|▌ | 85.2k/846k [00:00<00:01, 625kB/s]
sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.11M [00:00<00:06, 616kB
sub-NTNU1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.61M [00:00<?, ?B/s]
sub-NTNU1_acq-anat_TB1TFL.nii.gz: 48%|██▉ | 408k/846k [00:00<00:00, 1.64MB/s]
sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.11M [00:00<00:02, 1.43MB
sub-NTNU1_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.61M [00:00<00:07, 5
sub-NTNU1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz: 35%|▎| 1.46M/4.11M [00:00<00:00, 4.59M
sub-NTNU1_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.61M [00:00<00:02, 1.
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.6M [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz: 91%|▉| 3.73M/4.11M [00:00<00:00, 10.7M
sub-NTNU1_acq-coilQaSagSmall_GFactor.nii.gz: 33%|▎| 1.19M/3.61M [00:00<00:00, 4
sub-NTNU1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 0%| | 77.0k/15.6M [00:00<00:28, 567kB/
sub-NTNU1_acq-coilQaSagSmall_GFactor.nii.gz: 82%|▊| 2.95M/3.61M [00:00<00:00, 8
sub-NTNU1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.6k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 2%| | 366k/15.6M [00:00<00:10, 1.48MB/
sub-NTNU1_acq-famp-0.66_TB1DREAM.nii.gz: 94%|▉| 83.6k/88.6k [00:00<00:00, 619kB
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 9%| | 1.34M/15.6M [00:00<00:03, 4.53MB
sub-NTNU1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-NTNU1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/89.6k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 20%|▏| 3.16M/15.6M [00:00<00:01, 9.43MB
sub-NTNU1_acq-famp_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 43%|▍| 6.65M/15.6M [00:00<00:00, 18.2MB
sub-NTNU1_acq-famp_TB1DREAM.nii.gz: 86%|██▌| 77.0k/89.6k [00:00<00:00, 558kB/s]
sub-NTNU1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 62%|▌| 9.62M/15.6M [00:00<00:00, 21.9MB
sub-NTNU1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/951k [00:00<?, ?B/s]
sub-NTNU1_acq-coilQaTra_GFactor.nii.gz: 94%|▉| 14.7M/15.6M [00:00<00:00, 31.6MB
sub-NTNU1_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/951k [00:00<00:01, 604kB/s]
sub-NTNU1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/84.7k [00:00<?, ?B/s]
sub-NTNU1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU1_acq-famp_TB1TFL.nii.gz: 37%|██▏ | 356k/951k [00:00<00:00, 1.48MB/s]
sub-NTNU1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/89.3k [00:00<?, ?B/s]
sub-NTNU1_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 84.7k/84.7k [00:00<00:00, 621kB
sub-NTNU2_T2starw.json: 0%| | 0.00/2.54k [00:00<?, ?B/s]
sub-NTNU1_acq-refv_TB1DREAM.nii.gz: 76%|██▎| 67.6k/89.3k [00:00<00:00, 464kB/s]
sub-NTNU2_T2starw.nii.gz: 0%| | 0.00/1.32M [00:00<?, ?B/s]
sub-NTNU2_UNIT1.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU2_T2starw.nii.gz: 6%|▊ | 85.2k/1.32M [00:00<00:02, 591kB/s]
sub-NTNU2_inv-1_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU2_UNIT1.nii.gz: 0%| | 0.00/26.1M [00:00<?, ?B/s]
sub-NTNU2_T2starw.nii.gz: 27%|███▍ | 357k/1.32M [00:00<00:00, 1.44MB/s]
sub-NTNU2_UNIT1.nii.gz: 0%| | 77.0k/26.1M [00:00<00:41, 653kB/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.3M [00:00<?, ?B/s]
sub-NTNU2_inv-2_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU2_T2starw.nii.gz: 69%|█████████ | 935k/1.32M [00:00<00:00, 3.01MB/s]
sub-NTNU2_UNIT1.nii.gz: 1%|▏ | 366k/26.1M [00:00<00:17, 1.56MB/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 67.5k/18.3M [00:00<00:38, 495kB
sub-NTNU2_UNIT1.nii.gz: 6%|▊ | 1.45M/26.1M [00:00<00:04, 5.42MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/19.9M [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 339k/18.3M [00:00<00:13, 1.36MB
sub-NTNU2_UNIT1.nii.gz: 11%|█▌ | 2.95M/26.1M [00:00<00:02, 8.74MB/s]
sub-NTNU2_rec-uncombined01_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 4%| | 747k/18.3M [00:00<00:07, 2.41MB
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 85.2k/19.9M [00:00<00:33, 619kB
sub-NTNU2_UNIT1.nii.gz: 20%|██▊ | 5.14M/26.1M [00:00<00:01, 13.4MB/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.34M/18.3M [00:00<00:02, 7.42M
sub-NTNU2_rec-uncombined01_T2starw.nii.gz: 8%| | 83.6k/1.02M [00:00<00:01, 624
sub-NTNU2_UNIT1.nii.gz: 30%|████▏ | 7.85M/26.1M [00:00<00:01, 18.2MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 408k/19.9M [00:00<00:12, 1.64MB
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 24%|▏| 4.43M/18.3M [00:00<00:01, 12.2M
sub-NTNU2_UNIT1.nii.gz: 38%|█████▎ | 9.93M/26.1M [00:00<00:00, 19.3MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.13M/19.9M [00:00<00:05, 3.89M
sub-NTNU2_rec-uncombined01_T2starw.nii.gz: 34%|▎| 356k/1.02M [00:00<00:00, 1.43
sub-NTNU2_rec-uncombined02_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 33%|▎| 6.04M/18.3M [00:00<00:00, 13.3M
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 9%| | 1.74M/19.9M [00:00<00:03, 4.78M
sub-NTNU2_rec-uncombined01_T2starw.nii.gz: 94%|▉| 986k/1.02M [00:00<00:00, 3.31
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 40%|▍| 7.37M/18.3M [00:00<00:00, 12.8M
sub-NTNU2_UNIT1.nii.gz: 45%|██████▎ | 11.8M/26.1M [00:00<00:00, 15.8MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 13%|▏| 2.62M/19.9M [00:00<00:02, 6.22M
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 51%|▌| 9.29M/18.3M [00:00<00:00, 15.0M
sub-NTNU2_UNIT1.nii.gz: 53%|███████▎ | 13.7M/26.1M [00:01<00:00, 16.8MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 23%|▏| 4.52M/19.9M [00:00<00:01, 10.6M
sub-NTNU2_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/983k [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 61%|▌| 11.2M/18.3M [00:01<00:00, 16.4M
sub-NTNU2_UNIT1.nii.gz: 60%|████████▍ | 15.6M/26.1M [00:01<00:00, 17.7MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 32%|▎| 6.40M/19.9M [00:00<00:01, 13.4M
sub-NTNU2_rec-uncombined03_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 70%|▋| 12.8M/18.3M [00:01<00:00, 15.9M
sub-NTNU2_UNIT1.nii.gz: 67%|█████████▎ | 17.4M/26.1M [00:01<00:00, 17.0MB/s]
sub-NTNU2_rec-uncombined02_T2starw.nii.gz: 8%| | 83.6k/983k [00:00<00:01, 619k
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 39%|▍| 7.82M/19.9M [00:00<00:00, 13.8M
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 80%|▊| 14.6M/18.3M [00:01<00:00, 16.9M
sub-NTNU2_UNIT1.nii.gz: 74%|██████████▎ | 19.3M/26.1M [00:01<00:00, 17.7MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 48%|▍| 9.62M/19.9M [00:00<00:00, 15.3M
sub-NTNU2_rec-uncombined02_T2starw.nii.gz: 36%|▎| 356k/983k [00:00<00:00, 1.43M
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 89%|▉| 16.3M/18.3M [00:01<00:00, 17.1M
sub-NTNU2_UNIT1.nii.gz: 80%|███████████▎ | 21.0M/26.1M [00:01<00:00, 17.8MB/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 57%|▌| 11.4M/19.9M [00:01<00:00, 16.2M
sub-NTNU2_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz: 98%|▉| 18.0M/18.3M [00:01<00:00, 15.9M
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 65%|▋| 12.9M/19.9M [00:01<00:00, 15.3M
sub-NTNU2_UNIT1.nii.gz: 87%|████████████▏ | 22.8M/26.1M [00:01<00:00, 16.3MB/s]
sub-NTNU2_rec-uncombined03_T2starw.nii.gz: 9%| | 94.0k/1.01M [00:00<00:01, 864
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 75%|▊| 14.9M/19.9M [00:01<00:00, 16.8M
sub-NTNU2_UNIT1.nii.gz: 95%|█████████████▎| 24.7M/26.1M [00:01<00:00, 17.5MB/s]
sub-NTNU2_rec-uncombined03_T2starw.nii.gz: 34%|▎| 349k/1.01M [00:00<00:00, 1.57
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 85%|▊| 16.8M/19.9M [00:01<00:00, 17.7M
sub-NTNU2_rec-uncombined04_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_inv-2_part-mag_MP2RAGE.nii.gz: 95%|▉| 18.9M/19.9M [00:01<00:00, 19.1M
sub-NTNU2_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined05_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined04_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 570
sub-NTNU2_rec-uncombined06_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined04_T2starw.nii.gz: 33%|▎| 339k/1.00M [00:00<00:00, 1.36
sub-NTNU2_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined05_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 577
sub-NTNU2_rec-uncombined07_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined06_T2starw.nii.gz: 8%| | 83.5k/1.00M [00:00<00:01, 653
sub-NTNU2_rec-uncombined05_T2starw.nii.gz: 36%|▎| 356k/0.98M [00:00<00:00, 1.45
sub-NTNU2_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined06_T2starw.nii.gz: 40%|▍| 407k/1.00M [00:00<00:00, 1.67
sub-NTNU2_rec-uncombined07_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 590
sub-NTNU2_rec-uncombined09_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined08_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined07_T2starw.nii.gz: 33%|▎| 356k/1.05M [00:00<00:00, 1.44
sub-NTNU2_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/999k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined10_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined08_T2starw.nii.gz: 8%| | 83.6k/999k [00:00<00:01, 617k
sub-NTNU2_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined09_T2starw.nii.gz: 8%| | 85.2k/1.04M [00:00<00:01, 619
sub-NTNU2_rec-uncombined11_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined08_T2starw.nii.gz: 34%|▎| 339k/999k [00:00<00:00, 1.36M
sub-NTNU2_rec-uncombined10_T2starw.nii.gz: 6%| | 66.6k/1.04M [00:00<00:02, 489
sub-NTNU2_rec-uncombined09_T2starw.nii.gz: 34%|▎| 357k/1.04M [00:00<00:00, 1.42
sub-NTNU2_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/1.04M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined10_T2starw.nii.gz: 32%|▎| 339k/1.04M [00:00<00:00, 1.36
sub-NTNU2_rec-uncombined11_T2starw.nii.gz: 8%| | 83.6k/1.04M [00:00<00:01, 700
sub-NTNU2_rec-uncombined12_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined13_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined11_T2starw.nii.gz: 32%|▎| 339k/1.04M [00:00<00:00, 1.29
sub-NTNU2_rec-uncombined12_T2starw.nii.gz: 8%| | 83.5k/1.07M [00:00<00:01, 614
sub-NTNU2_rec-uncombined14_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/1.05M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined12_T2starw.nii.gz: 37%|▎| 407k/1.07M [00:00<00:00, 1.65
sub-NTNU2_rec-uncombined15_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined13_T2starw.nii.gz: 8%| | 83.6k/1.05M [00:00<00:01, 677
sub-NTNU2_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.10M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined13_T2starw.nii.gz: 36%|▎| 390k/1.05M [00:00<00:00, 1.60
sub-NTNU2_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined14_T2starw.nii.gz: 7%| | 83.6k/1.10M [00:00<00:01, 610
sub-NTNU2_rec-uncombined16_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined15_T2starw.nii.gz: 8%| | 83.6k/1.06M [00:00<00:01, 527
sub-NTNU2_rec-uncombined14_T2starw.nii.gz: 32%|▎| 356k/1.10M [00:00<00:00, 1.41
sub-NTNU2_rec-uncombined15_T2starw.nii.gz: 31%|▎| 339k/1.06M [00:00<00:00, 1.39
sub-NTNU2_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/1.03M [00:00<00:01, 616
sub-NTNU2_rec-uncombined17_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined15_T2starw.nii.gz: 70%|▋| 764k/1.06M [00:00<00:00, 2.49
sub-NTNU2_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/993k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined16_T2starw.nii.gz: 32%|▎| 339k/1.03M [00:00<00:00, 1.36
sub-NTNU2_rec-uncombined18_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined17_T2starw.nii.gz: 7%| | 66.6k/993k [00:00<00:02, 442k
sub-NTNU2_rec-uncombined17_T2starw.nii.gz: 32%|▎| 322k/993k [00:00<00:00, 1.32M
sub-NTNU2_rec-uncombined19_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/973k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined20_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined18_T2starw.nii.gz: 9%| | 83.6k/973k [00:00<00:01, 653k
sub-NTNU2_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined18_T2starw.nii.gz: 37%|▎| 356k/973k [00:00<00:00, 1.46M
sub-NTNU2_rec-uncombined19_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 613
sub-NTNU2_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/987k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined21_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined18_T2starw.nii.gz: 99%|▉| 968k/973k [00:00<00:00, 3.18M
sub-NTNU2_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/980k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined19_T2starw.nii.gz: 38%|▍| 390k/0.99M [00:00<00:00, 1.57
sub-NTNU2_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/987k [00:00<00:01, 610k
sub-NTNU2_rec-uncombined21_T2starw.nii.gz: 9%| | 83.6k/980k [00:00<00:01, 695k
sub-NTNU2_rec-uncombined20_T2starw.nii.gz: 26%|▎| 254k/987k [00:00<00:00, 1.18M
sub-NTNU2_rec-uncombined21_T2starw.nii.gz: 35%|▎| 339k/980k [00:00<00:00, 1.42M
sub-NTNU2_rec-uncombined20_T2starw.nii.gz: 72%|▋| 713k/987k [00:00<00:00, 2.55M
sub-NTNU2_rec-uncombined22_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined23_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined22_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 787
sub-NTNU2_rec-uncombined22_T2starw.nii.gz: 22%|▏| 220k/0.98M [00:00<00:00, 1.10
sub-NTNU2_rec-uncombined24_T2starw.json: 0%| | 0.00/2.55k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/991k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined22_T2starw.nii.gz: 90%|▉| 900k/0.98M [00:00<00:00, 3.44
sub-NTNU2_rec-uncombined23_T2starw.nii.gz: 8%| | 83.6k/991k [00:00<00:01, 664k
sub-NTNU2_acq-anat_TB1TFL.json: 0%| | 0.00/2.68k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined24_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 619
sub-NTNU2_rec-uncombined23_T2starw.nii.gz: 36%|▎| 356k/991k [00:00<00:00, 1.58M
sub-NTNU2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.28k [00:00<?, ?B/s]
sub-NTNU2_rec-uncombined24_T2starw.nii.gz: 33%|▎| 339k/1.00M [00:00<00:00, 1.63
sub-NTNU2_rec-uncombined24_T2starw.nii.gz: 73%|▋| 747k/1.00M [00:00<00:00, 2.60
sub-NTNU2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/731k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.04M [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.28k [00:00<?, ?B/s]
sub-NTNU2_acq-anat_TB1TFL.nii.gz: 12%|▋ | 85.2k/731k [00:00<00:01, 627kB/s]
sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.04M [00:00<00:06, 612kB
sub-NTNU2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.62M [00:00<?, ?B/s]
sub-NTNU2_acq-anat_TB1TFL.nii.gz: 49%|██▉ | 357k/731k [00:00<00:00, 1.43MB/s]
sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz: 9%| | 373k/4.04M [00:00<00:02, 1.49MB
sub-NTNU2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.25k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz: 20%|▏| 815k/4.04M [00:00<00:01, 2.54MB
sub-NTNU2_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.62M [00:00<00:06, 5
sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz: 76%|▊| 3.09M/4.04M [00:00<00:00, 9.85M
sub-NTNU2_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.62M [00:00<00:02, 1.
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.5M [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagSmall_GFactor.nii.gz: 40%|▍| 1.46M/3.62M [00:00<00:00, 4
sub-NTNU2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.6k [00:00<?, ?B/s]
sub-NTNU2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.25k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaSagSmall_GFactor.nii.gz: 84%|▊| 3.03M/3.62M [00:00<00:00, 8
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 0%| | 77.0k/15.5M [00:00<00:28, 562kB/
sub-NTNU2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.6k [00:00<?, ?B/s]
sub-NTNU2_acq-famp-0.66_TB1DREAM.nii.gz: 94%|▉| 83.6k/88.6k [00:00<00:00, 614kB
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 2%| | 383k/15.5M [00:00<00:10, 1.55MB/
sub-NTNU2_acq-famp-1.5_TB1DREAM.nii.gz: 74%|▋| 66.6k/89.6k [00:00<00:00, 488kB/
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 7%| | 1.04M/15.5M [00:00<00:04, 3.51MB
sub-NTNU2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.24k [00:00<?, ?B/s]
sub-NTNU2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/89.1k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 20%|▏| 3.13M/15.5M [00:00<00:01, 9.64MB
sub-NTNU2_acq-famp_TB1TFL.json: 0%| | 0.00/2.72k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 42%|▍| 6.47M/15.5M [00:00<00:00, 17.9MB
sub-NTNU2_acq-famp_TB1DREAM.nii.gz: 94%|██▊| 83.5k/89.1k [00:00<00:00, 614kB/s]
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 68%|▋| 10.5M/15.5M [00:00<00:00, 25.5MB
sub-NTNU2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/935k [00:00<?, ?B/s]
sub-NTNU2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.27k [00:00<?, ?B/s]
sub-NTNU2_acq-coilQaTra_GFactor.nii.gz: 89%|▉| 13.8M/15.5M [00:00<00:00, 28.3MB
sub-NTNU2_acq-famp_TB1TFL.nii.gz: 10%|▌ | 94.0k/935k [00:00<00:01, 685kB/s]
sub-NTNU2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/82.6k [00:00<?, ?B/s]
sub-NTNU2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.27k [00:00<?, ?B/s]
sub-NTNU2_acq-famp_TB1TFL.nii.gz: 43%|██▌ | 400k/935k [00:00<00:00, 1.58MB/s]
sub-NTNU2_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 82.6k/82.6k [00:00<00:00, 602kB
sub-NTNU2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.26k [00:00<?, ?B/s]
sub-NTNU2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/89.8k [00:00<?, ?B/s]
sub-NTNU2_acq-refv-1.5_TB1DREAM.nii.gz: 93%|▉| 83.6k/89.8k [00:00<00:00, 653kB/
sub-NTNU2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/85.9k [00:00<?, ?B/s]
sub-NTNU3_T2starw.nii.gz: 0%| | 0.00/1.33M [00:00<?, ?B/s]
sub-NTNU3_UNIT1.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU2_acq-refv_TB1DREAM.nii.gz: 99%|██▉| 85.2k/85.9k [00:00<00:00, 544kB/s]
sub-NTNU3_T2starw.json: 0%| | 0.00/2.47k [00:00<?, ?B/s]
sub-NTNU3_T2starw.nii.gz: 4%|▍ | 52.2k/1.33M [00:00<00:02, 469kB/s]
sub-NTNU3_T2starw.nii.gz: 19%|██▍ | 255k/1.33M [00:00<00:00, 1.33MB/s]
sub-NTNU3_UNIT1.nii.gz: 0%| | 0.00/25.8M [00:00<?, ?B/s]
sub-NTNU3_inv-1_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU3_T2starw.nii.gz: 54%|██████▉ | 731k/1.33M [00:00<00:00, 2.52MB/s]
sub-NTNU3_inv-2_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-NTNU3_UNIT1.nii.gz: 0%| | 83.5k/25.8M [00:00<00:45, 590kB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/18.6M [00:00<?, ?B/s]
sub-NTNU3_UNIT1.nii.gz: 1%|▏ | 390k/25.8M [00:00<00:14, 1.80MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/18.6M [00:00<00:31, 619kB
sub-NTNU3_UNIT1.nii.gz: 3%|▍ | 815k/25.8M [00:00<00:09, 2.84MB/s]
sub-NTNU3_rec-uncombined01_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_UNIT1.nii.gz: 7%|▉ | 1.84M/25.8M [00:00<00:04, 5.77MB/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/20.4M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined01_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 356k/18.6M [00:00<00:13, 1.43MB
sub-NTNU3_UNIT1.nii.gz: 22%|███ | 5.58M/25.8M [00:00<00:01, 17.2MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 6%| | 1.19M/18.6M [00:00<00:04, 4.23M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/20.4M [00:00<00:30, 711kB
sub-NTNU3_rec-uncombined01_T2starw.nii.gz: 8%| | 83.5k/1.06M [00:00<00:01, 611
sub-NTNU3_UNIT1.nii.gz: 34%|████▋ | 8.68M/25.8M [00:00<00:00, 22.1MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 14%|▏| 2.68M/18.6M [00:00<00:02, 7.93M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 356k/20.4M [00:00<00:13, 1.51MB
sub-NTNU3_rec-uncombined02_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined01_T2starw.nii.gz: 36%|▎| 391k/1.06M [00:00<00:00, 1.56
sub-NTNU3_UNIT1.nii.gz: 43%|██████ | 11.1M/25.8M [00:00<00:00, 23.2MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 25%|▏| 4.60M/18.6M [00:00<00:01, 11.9M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 5%| | 1.01M/20.4M [00:00<00:05, 3.63M
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 31%|▎| 5.82M/18.6M [00:00<00:01, 12.1M
sub-NTNU3_rec-uncombined02_T2starw.nii.gz: 8%| | 77.0k/1.00M [00:00<00:01, 564
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 8%| | 1.73M/20.4M [00:00<00:03, 5.00M
sub-NTNU3_UNIT1.nii.gz: 52%|███████▎ | 13.4M/25.8M [00:00<00:00, 19.6MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 41%|▍| 7.53M/18.6M [00:00<00:00, 13.9M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 17%|▏| 3.50M/20.4M [00:00<00:01, 9.57M
sub-NTNU3_rec-uncombined02_T2starw.nii.gz: 36%|▎| 366k/1.00M [00:00<00:00, 1.46
sub-NTNU3_UNIT1.nii.gz: 60%|████████▎ | 15.4M/25.8M [00:01<00:00, 19.3MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 50%|▍| 9.20M/18.6M [00:00<00:00, 15.0M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 25%|▏| 5.09M/20.4M [00:00<00:01, 11.8M
sub-NTNU3_rec-uncombined02_T2starw.nii.gz: 99%|▉| 0.99M/1.00M [00:00<00:00, 3.3
sub-NTNU3_rec-uncombined02_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_UNIT1.nii.gz: 67%|█████████▍ | 17.3M/25.8M [00:01<00:00, 16.3MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 57%|▌| 10.7M/18.6M [00:01<00:00, 13.6M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 31%|▎| 6.26M/20.4M [00:00<00:01, 11.5M
sub-NTNU3_UNIT1.nii.gz: 74%|██████████▍ | 19.2M/25.8M [00:01<00:00, 17.2MB/s]
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 68%|▋| 12.5M/18.6M [00:01<00:00, 15.3M
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 40%|▍| 8.15M/20.4M [00:00<00:00, 14.0M
sub-NTNU3_UNIT1.nii.gz: 82%|███████████▍ | 21.1M/25.8M [00:01<00:00, 17.9MB/s]
sub-NTNU3_rec-uncombined04_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 47%|▍| 9.65M/20.4M [00:00<00:00, 14.2M
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 78%|▊| 14.5M/18.6M [00:01<00:00, 15.6M
sub-NTNU3_rec-uncombined03_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 54%|▌| 11.1M/20.4M [00:01<00:00, 14.4M
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 86%|▊| 16.0M/18.6M [00:01<00:00, 15.5M
sub-NTNU3_UNIT1.nii.gz: 89%|████████████▍ | 22.9M/25.8M [00:01<00:00, 16.2MB/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 64%|▋| 13.1M/20.4M [00:01<00:00, 16.3M
sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz: 97%|▉| 17.9M/18.6M [00:01<00:00, 16.8M
sub-NTNU3_UNIT1.nii.gz: 96%|█████████████▍| 24.8M/25.8M [00:01<00:00, 17.2MB/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 72%|▋| 14.7M/20.4M [00:01<00:00, 16.0M
sub-NTNU3_rec-uncombined03_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined05_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_inv-2_part-mag_MP2RAGE.nii.gz: 91%|▉| 18.5M/20.4M [00:01<00:00, 23.1M
sub-NTNU3_rec-uncombined03_T2starw.nii.gz: 8%| | 83.5k/1.03M [00:00<00:01, 611
sub-NTNU3_rec-uncombined04_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined03_T2starw.nii.gz: 38%|▍| 407k/1.03M [00:00<00:00, 1.59
sub-NTNU3_rec-uncombined05_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined06_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined04_T2starw.nii.gz: 8%| | 78.0k/1.01M [00:00<00:01, 574
sub-NTNU3_rec-uncombined05_T2starw.nii.gz: 9%| | 94.0k/1.02M [00:00<00:01, 691
sub-NTNU3_rec-uncombined06_T2starw.nii.gz: 0%| | 0.00/1.03M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined05_T2starw.nii.gz: 29%|▎| 298k/1.02M [00:00<00:00, 1.40
sub-NTNU3_rec-uncombined04_T2starw.nii.gz: 35%|▎| 366k/1.01M [00:00<00:00, 1.47
sub-NTNU3_rec-uncombined06_T2starw.nii.gz: 8%| | 83.5k/1.03M [00:00<00:01, 613
sub-NTNU3_rec-uncombined07_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined05_T2starw.nii.gz: 69%|▋| 723k/1.02M [00:00<00:00, 2.49
sub-NTNU3_rec-uncombined04_T2starw.nii.gz: 78%|▊| 809k/1.01M [00:00<00:00, 2.62
sub-NTNU3_rec-uncombined06_T2starw.nii.gz: 29%|▎| 305k/1.03M [00:00<00:00, 1.41
sub-NTNU3_rec-uncombined07_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined06_T2starw.nii.gz: 79%|▊| 832k/1.03M [00:00<00:00, 2.97
sub-NTNU3_rec-uncombined07_T2starw.nii.gz: 8%| | 83.5k/1.08M [00:00<00:01, 622
sub-NTNU3_rec-uncombined08_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined09_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined07_T2starw.nii.gz: 37%|▎| 407k/1.08M [00:00<00:00, 1.54
sub-NTNU3_rec-uncombined08_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined10_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined08_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 617
sub-NTNU3_rec-uncombined08_T2starw.nii.gz: 35%|▎| 356k/0.99M [00:00<00:00, 1.43
sub-NTNU3_rec-uncombined10_T2starw.nii.gz: 0%| | 0.00/1.07M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined09_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined12_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined10_T2starw.nii.gz: 8%| | 83.6k/1.07M [00:00<00:01, 611
sub-NTNU3_rec-uncombined11_T2starw.nii.gz: 0%| | 0.00/1.11M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined09_T2starw.nii.gz: 6%| | 66.6k/1.08M [00:00<00:01, 607
sub-NTNU3_rec-uncombined10_T2starw.nii.gz: 32%|▎| 356k/1.07M [00:00<00:00, 1.41
sub-NTNU3_rec-uncombined09_T2starw.nii.gz: 29%|▎| 322k/1.08M [00:00<00:00, 1.49
sub-NTNU3_rec-uncombined11_T2starw.nii.gz: 7%| | 83.6k/1.11M [00:00<00:01, 614
sub-NTNU3_rec-uncombined09_T2starw.nii.gz: 72%|▋| 798k/1.08M [00:00<00:00, 2.70
sub-NTNU3_rec-uncombined11_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined11_T2starw.nii.gz: 34%|▎| 390k/1.11M [00:00<00:00, 1.55
sub-NTNU3_rec-uncombined12_T2starw.nii.gz: 0%| | 0.00/1.10M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined12_T2starw.nii.gz: 7%| | 83.6k/1.10M [00:00<00:01, 609
sub-NTNU3_rec-uncombined13_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined12_T2starw.nii.gz: 31%|▎| 356k/1.10M [00:00<00:00, 1.42
sub-NTNU3_rec-uncombined13_T2starw.nii.gz: 0%| | 0.00/1.08M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined14_T2starw.nii.gz: 0%| | 0.00/1.12M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined14_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined12_T2starw.nii.gz: 89%|▉| 0.98M/1.10M [00:00<00:00, 3.3
sub-NTNU3_rec-uncombined13_T2starw.nii.gz: 8%| | 83.6k/1.08M [00:00<00:01, 613
sub-NTNU3_rec-uncombined14_T2starw.nii.gz: 6%| | 66.6k/1.12M [00:00<00:02, 487
sub-NTNU3_rec-uncombined15_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined14_T2starw.nii.gz: 18%|▏| 204k/1.12M [00:00<00:01, 944k
sub-NTNU3_rec-uncombined13_T2starw.nii.gz: 23%|▏| 254k/1.08M [00:00<00:00, 1.14
sub-NTNU3_rec-uncombined13_T2starw.nii.gz: 66%|▋| 730k/1.08M [00:00<00:00, 2.65
sub-NTNU3_rec-uncombined16_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined15_T2starw.nii.gz: 0%| | 0.00/1.09M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined14_T2starw.nii.gz: 58%|▌| 662k/1.12M [00:00<00:00, 2.19
sub-NTNU3_rec-uncombined15_T2starw.nii.gz: 8%| | 85.2k/1.09M [00:00<00:01, 737
sub-NTNU3_rec-uncombined16_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined15_T2starw.nii.gz: 31%|▎| 340k/1.09M [00:00<00:00, 1.46
sub-NTNU3_rec-uncombined17_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined16_T2starw.nii.gz: 8%| | 83.6k/1.06M [00:00<00:01, 616
sub-NTNU3_rec-uncombined18_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined17_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined15_T2starw.nii.gz: 75%|▋| 833k/1.09M [00:00<00:00, 2.86
sub-NTNU3_rec-uncombined17_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 610
sub-NTNU3_rec-uncombined16_T2starw.nii.gz: 33%|▎| 356k/1.06M [00:00<00:00, 1.42
sub-NTNU3_rec-uncombined17_T2starw.nii.gz: 30%|▎| 305k/0.99M [00:00<00:00, 1.19
sub-NTNU3_rec-uncombined20_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined18_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined19_T2starw.nii.gz: 0%| | 0.00/1.01M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined19_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined18_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 650
sub-NTNU3_rec-uncombined19_T2starw.nii.gz: 8%| | 83.6k/1.01M [00:00<00:01, 622
sub-NTNU3_rec-uncombined18_T2starw.nii.gz: 35%|▎| 356k/0.98M [00:00<00:00, 1.46
sub-NTNU3_rec-uncombined21_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined19_T2starw.nii.gz: 34%|▎| 356k/1.01M [00:00<00:00, 1.40
sub-NTNU3_rec-uncombined20_T2starw.nii.gz: 0%| | 0.00/0.99M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined22_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined20_T2starw.nii.gz: 8%| | 83.6k/0.99M [00:00<00:01, 644
sub-NTNU3_rec-uncombined20_T2starw.nii.gz: 35%|▎| 356k/0.99M [00:00<00:00, 1.54
sub-NTNU3_rec-uncombined22_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined22_T2starw.nii.gz: 8%| | 85.2k/1.00M [00:00<00:01, 526
sub-NTNU3_rec-uncombined21_T2starw.nii.gz: 0%| | 0.00/0.98M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined23_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined20_T2starw.nii.gz: 79%|▊| 798k/0.99M [00:00<00:00, 2.70
sub-NTNU3_rec-uncombined22_T2starw.nii.gz: 40%|▍| 408k/1.00M [00:00<00:00, 1.67
sub-NTNU3_rec-uncombined21_T2starw.nii.gz: 8%| | 83.6k/0.98M [00:00<00:01, 653
sub-NTNU3_rec-uncombined21_T2starw.nii.gz: 35%|▎| 356k/0.98M [00:00<00:00, 1.45
sub-NTNU3_rec-uncombined23_T2starw.nii.gz: 0%| | 0.00/994k [00:00<?, ?B/s]
sub-NTNU3_acq-anat_TB1TFL.json: 0%| | 0.00/2.70k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined24_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined23_T2starw.nii.gz: 5%| | 49.6k/994k [00:00<00:01, 503k
sub-NTNU3_rec-uncombined23_T2starw.nii.gz: 17%|▏| 169k/994k [00:00<00:00, 880kB
sub-NTNU3_rec-uncombined24_T2starw.nii.gz: 0%| | 0.00/1.00M [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined23_T2starw.nii.gz: 73%|▋| 730k/994k [00:00<00:00, 2.58M
sub-NTNU3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/834k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.21k [00:00<?, ?B/s]
sub-NTNU3_rec-uncombined24_T2starw.nii.gz: 8%| | 83.6k/1.00M [00:00<00:01, 566
sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.09M [00:00<?, ?B/s]
sub-NTNU3_acq-anat_TB1TFL.nii.gz: 10%|▌ | 83.6k/834k [00:00<00:01, 613kB/s]
sub-NTNU3_rec-uncombined24_T2starw.nii.gz: 38%|▍| 390k/1.00M [00:00<00:00, 1.59
sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.09M [00:00<00:07, 580kB
sub-NTNU3_acq-anat_TB1TFL.nii.gz: 41%|██▍ | 339k/834k [00:00<00:00, 1.35MB/s]
sub-NTNU3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.23k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.09M [00:00<00:02, 1.42MB
sub-NTNU3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/3.62M [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz: 36%|▎| 1.46M/4.09M [00:00<00:00, 4.56M
sub-NTNU3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagSmall_GFactor.nii.gz: 2%| | 83.6k/3.62M [00:00<00:07, 5
sub-NTNU3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz: 89%|▉| 3.64M/4.09M [00:00<00:00, 10.1M
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/15.6M [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 356k/3.62M [00:00<00:02, 1.
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 0%| | 66.6k/15.6M [00:00<00:26, 611kB/
sub-NTNU3_acq-coilQaSagSmall_GFactor.nii.gz: 41%|▍| 1.48M/3.62M [00:00<00:00, 4
sub-NTNU3_acq-famp_TB1TFL.json: 0%| | 0.00/2.73k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaSagSmall_GFactor.nii.gz: 100%|▉| 3.60M/3.62M [00:00<00:00, 1
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 2%| | 339k/15.6M [00:00<00:11, 1.35MB/
sub-NTNU3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-NTNU3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/88.1k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 9%| | 1.38M/15.6M [00:00<00:03, 4.65MB
sub-NTNU3_acq-famp-0.66_TB1DREAM.nii.gz: 97%|▉| 85.2k/88.1k [00:00<00:00, 633kB
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 20%|▏| 3.06M/15.6M [00:00<00:01, 8.99MB
sub-NTNU3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/89.3k [00:00<?, ?B/s]
sub-NTNU3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 46%|▍| 7.23M/15.6M [00:00<00:00, 19.0MB
sub-NTNU3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/955k [00:00<?, ?B/s]
sub-NTNU3_acq-coilQaTra_GFactor.nii.gz: 76%|▊| 11.9M/15.6M [00:00<00:00, 27.8MB
sub-NTNU3_acq-famp_TB1DREAM.nii.gz: 94%|██▊| 83.6k/89.3k [00:00<00:00, 618kB/s]
sub-NTNU3_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/955k [00:00<00:01, 610kB/s]
sub-NTNU3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/84.4k [00:00<?, ?B/s]
sub-NTNU3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-NTNU3_acq-famp_TB1TFL.nii.gz: 37%|██▏ | 356k/955k [00:00<00:00, 1.42MB/s]
sub-NTNU3_acq-refv-0.66_TB1DREAM.nii.gz: 99%|▉| 83.6k/84.4k [00:00<00:00, 617kB
sub-NTNU3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/89.6k [00:00<?, ?B/s]
sub-UCL1_T2starw.json: 0%| | 0.00/2.48k [00:00<?, ?B/s]
sub-UCL1_UNIT1.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-NTNU3_acq-refv_TB1DREAM.nii.gz: 86%|██▌| 77.0k/89.6k [00:00<00:00, 561kB/s]
sub-UCL1_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 0%| | 0.00/26.5M [00:00<?, ?B/s]
sub-UCL1_inv-1_MP2RAGE.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL1_T2starw.nii.gz: 8%|█ | 85.2k/1.06M [00:00<00:01, 579kB/s]
sub-UCL1_inv-2_MP2RAGE.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 0%| | 83.6k/26.5M [00:00<00:45, 608kB/s]
sub-UCL1_T2starw.nii.gz: 33%|████▋ | 357k/1.06M [00:00<00:00, 1.56MB/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/15.4M [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 1%|▏ | 390k/26.5M [00:00<00:17, 1.56MB/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/15.4M [00:00<00:28, 565kB/
sub-UCL1_rec-uncombined1_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 6%|▊ | 1.54M/26.5M [00:00<00:05, 4.45MB/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.0M [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 18%|██▊ | 4.90M/26.5M [00:00<00:01, 13.7MB/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 2%| | 349k/15.4M [00:00<00:11, 1.40MB/
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 83.6k/17.0M [00:00<00:23, 747kB/
sub-UCL1_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/889k [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 30%|████▍ | 7.90M/26.5M [00:00<00:01, 19.0MB/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 7%| | 1.12M/15.4M [00:00<00:03, 3.94MB
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 390k/17.0M [00:00<00:10, 1.69MB/
sub-UCL1_UNIT1.nii.gz: 41%|██████ | 10.8M/26.5M [00:00<00:00, 22.3MB/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 2.52M/15.4M [00:00<00:01, 7.68MB
sub-UCL1_rec-uncombined1_T2starw.nii.gz: 7%| | 66.6k/889k [00:00<00:01, 485kB/
sub-UCL1_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/883k [00:00<?, ?B/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 1.08M/17.0M [00:00<00:04, 3.90MB
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 28%|▎| 4.35M/15.4M [00:00<00:01, 11.5MB
sub-UCL1_UNIT1.nii.gz: 50%|███████▍ | 13.2M/26.5M [00:00<00:00, 21.4MB/s]
sub-UCL1_rec-uncombined1_T2starw.nii.gz: 36%|▎| 322k/889k [00:00<00:00, 1.28MB/
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 12%| | 2.12M/17.0M [00:00<00:02, 6.41MB
sub-UCL1_rec-uncombined2_T2starw.nii.gz: 9%| | 83.5k/883k [00:00<00:01, 609kB/
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 39%|▍| 6.04M/15.4M [00:00<00:00, 13.5MB
sub-UCL1_rec-uncombined1_T2starw.nii.gz: 97%|▉| 866k/889k [00:00<00:00, 2.86MB/
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 21%|▏| 3.59M/17.0M [00:00<00:01, 9.43MB
sub-UCL1_UNIT1.nii.gz: 58%|████████▋ | 15.4M/26.5M [00:01<00:00, 18.4MB/s]
sub-UCL1_rec-uncombined2_T2starw.nii.gz: 40%|▍| 356k/883k [00:00<00:00, 1.42MB/
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 48%|▍| 7.40M/15.4M [00:00<00:00, 13.1MB
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 29%|▎| 4.89M/17.0M [00:00<00:01, 10.8MB
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 57%|▌| 8.69M/15.4M [00:00<00:00, 13.2MB
sub-UCL1_UNIT1.nii.gz: 65%|█████████▊ | 17.3M/26.5M [00:01<00:00, 17.1MB/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 37%|▎| 6.21M/17.0M [00:00<00:00, 11.8MB
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 69%|▋| 10.6M/15.4M [00:00<00:00, 15.3MB
sub-UCL1_UNIT1.nii.gz: 73%|██████████▉ | 19.2M/26.5M [00:01<00:00, 17.9MB/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 48%|▍| 8.23M/17.0M [00:00<00:00, 14.7MB
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 81%|▊| 12.4M/15.4M [00:01<00:00, 16.2MB
sub-UCL1_rec-uncombined2_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_UNIT1.nii.gz: 79%|███████████▉ | 21.0M/26.5M [00:01<00:00, 17.1MB/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 57%|▌| 9.65M/17.0M [00:00<00:00, 14.3MB
sub-UCL1_rec-uncombined3_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz: 91%|▉| 14.0M/15.4M [00:01<00:00, 14.8MB
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 65%|▋| 11.1M/17.0M [00:01<00:00, 14.6MB
sub-UCL1_UNIT1.nii.gz: 86%|████████████▊ | 22.7M/26.5M [00:01<00:00, 16.6MB/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 74%|▋| 12.6M/17.0M [00:01<00:00, 14.9MB
sub-UCL1_UNIT1.nii.gz: 92%|█████████████▊ | 24.4M/26.5M [00:01<00:00, 16.7MB/s]
sub-UCL1_rec-uncombined4_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_inv-2_part-mag_MP2RAGE.nii.gz: 87%|▊| 14.8M/17.0M [00:01<00:00, 17.3MB
sub-UCL1_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/871k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined3_T2starw.nii.gz: 10%| | 85.2k/871k [00:00<00:01, 625kB/
sub-UCL1_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/810k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined5_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/897k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined3_T2starw.nii.gz: 47%|▍| 408k/871k [00:00<00:00, 1.65MB/
sub-UCL1_rec-uncombined4_T2starw.nii.gz: 12%| | 94.0k/810k [00:00<00:00, 735kB/
sub-UCL1_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/862k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined5_T2starw.nii.gz: 9%| | 83.6k/897k [00:00<00:01, 613kB/
sub-UCL1_rec-uncombined4_T2starw.nii.gz: 49%|▍| 400k/810k [00:00<00:00, 1.71MB/
sub-UCL1_rec-uncombined6_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined5_T2starw.nii.gz: 36%|▎| 323k/897k [00:00<00:00, 1.54MB/
sub-UCL1_rec-uncombined6_T2starw.nii.gz: 10%| | 85.2k/862k [00:00<00:01, 663kB/
sub-UCL1_rec-uncombined5_T2starw.nii.gz: 87%|▊| 781k/897k [00:00<00:00, 2.74MB/
sub-UCL1_rec-uncombined6_T2starw.nii.gz: 47%|▍| 408k/862k [00:00<00:00, 1.68MB/
sub-UCL1_rec-uncombined7_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined8_T2starw.json: 0%| | 0.00/2.49k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/828k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined7_T2starw.nii.gz: 10%| | 83.6k/828k [00:00<00:01, 661kB/
sub-UCL1_acq-anat_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/855k [00:00<?, ?B/s]
sub-UCL1_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/886k [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.19k [00:00<?, ?B/s]
sub-UCL1_rec-uncombined7_T2starw.nii.gz: 43%|▍| 356k/828k [00:00<00:00, 1.43MB/
sub-UCL1_rec-uncombined8_T2starw.nii.gz: 8%| | 67.5k/855k [00:00<00:01, 448kB/
sub-UCL1_acq-anat_TB1TFL.nii.gz: 9%|▋ | 83.6k/886k [00:00<00:01, 641kB/s]
sub-UCL1_rec-uncombined8_T2starw.nii.gz: 40%|▍| 339k/855k [00:00<00:00, 1.41MB/
sub-UCL1_acq-anat_TB1TFL.nii.gz: 40%|██▊ | 356k/886k [00:00<00:00, 1.50MB/s]
sub-UCL1_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-UCL1_acq-anat_TB1TFL.nii.gz: 82%|█████▊ | 730k/886k [00:00<00:00, 2.35MB/s]
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.19M [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.69M [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.6k/4.19M [00:00<00:07, 610kB/
sub-UCL1_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.69M [00:00<00:04, 61
sub-UCL1_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.10k [00:00<?, ?B/s]
sub-UCL1_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 5%| | 203k/4.19M [00:00<00:04, 860kB/s
sub-UCL1_acq-coilQaSagSmall_GFactor.nii.gz: 10%| | 288k/2.69M [00:00<00:01, 1.3
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 19%|▏| 798k/4.19M [00:00<00:01, 2.97MB/
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagSmall_GFactor.nii.gz: 26%|▎| 713k/2.69M [00:00<00:00, 2.4
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 55%|▌| 2.31M/4.19M [00:00<00:00, 7.73MB
sub-UCL1_acq-coilQaSagSmall_GFactor.nii.gz: 73%|▋| 1.98M/2.69M [00:00<00:00, 6.
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 1%| | 66.6k/11.7M [00:00<00:27, 451kB/s
sub-UCL1_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL1_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/93.0k [00:00<?, ?B/s]
sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz: 83%|▊| 3.48M/4.19M [00:00<00:00, 8.88MB
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 3%| | 322k/11.7M [00:00<00:08, 1.41MB/s
sub-UCL1_acq-famp-0.66_TB1DREAM.nii.gz: 84%|▊| 78.0k/93.0k [00:00<00:00, 607kB/
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 10%| | 1.18M/11.7M [00:00<00:02, 4.42MB/
sub-UCL1_acq-famp_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 22%|▏| 2.60M/11.7M [00:00<00:01, 7.80MB/
sub-UCL1_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.9k [00:00<?, ?B/s]
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 55%|▌| 6.41M/11.7M [00:00<00:00, 18.4MB/
sub-UCL1_acq-famp_TB1TFL.json: 0%| | 0.00/2.70k [00:00<?, ?B/s]
sub-UCL1_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/93.1k [00:00<?, ?B/s]
sub-UCL1_acq-famp-1.5_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.9k [00:00<00:00, 613kB/s
sub-UCL1_acq-coilQaTra_GFactor.nii.gz: 75%|▊| 8.82M/11.7M [00:00<00:00, 20.5MB/
sub-UCL1_acq-famp_TB1DREAM.nii.gz: 92%|███▋| 85.2k/93.1k [00:00<00:00, 754kB/s]
sub-UCL1_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/943k [00:00<?, ?B/s]
sub-UCL1_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL1_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL1_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/943k [00:00<00:01, 526kB/s]
sub-UCL1_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/86.7k [00:00<?, ?B/s]
sub-UCL1_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-UCL1_acq-famp_TB1TFL.nii.gz: 38%|██▋ | 356k/943k [00:00<00:00, 1.47MB/s]
sub-UCL1_acq-refv_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL1_acq-refv-0.66_TB1DREAM.nii.gz: 96%|▉| 83.6k/86.7k [00:00<00:00, 616kB/
sub-UCL1_acq-refv-1.5_TB1DREAM.nii.gz: 91%|▉| 83.6k/92.1k [00:00<00:00, 558kB/s
sub-UCL2_T2starw.nii.gz: 0%| | 0.00/1.02M [00:00<?, ?B/s]
sub-UCL2_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-UCL2_T2starw.nii.gz: 8%|█ | 83.6k/1.02M [00:00<00:01, 549kB/s]
sub-UCL2_UNIT1.json: 0%| | 0.00/2.08k [00:00<?, ?B/s]
sub-UCL1_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/89.4k [00:00<?, ?B/s]
sub-UCL2_T2starw.nii.gz: 34%|████▊ | 356k/1.02M [00:00<00:00, 1.39MB/s]
sub-UCL2_UNIT1.nii.gz: 0%| | 0.00/26.7M [00:00<?, ?B/s]
sub-UCL1_acq-refv_TB1DREAM.nii.gz: 93%|███▋| 83.6k/89.4k [00:00<00:00, 649kB/s]
sub-UCL2_inv-2_MP2RAGE.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-UCL2_UNIT1.nii.gz: 0%| | 83.6k/26.7M [00:00<00:45, 614kB/s]
sub-UCL2_inv-1_MP2RAGE.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-UCL2_UNIT1.nii.gz: 1%|▏ | 322k/26.7M [00:00<00:18, 1.54MB/s]
sub-UCL2_UNIT1.nii.gz: 3%|▍ | 730k/26.7M [00:00<00:10, 2.53MB/s]
sub-UCL2_rec-uncombined1_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/13.6M [00:00<?, ?B/s]
sub-UCL2_UNIT1.nii.gz: 9%|█▍ | 2.46M/26.7M [00:00<00:03, 8.28MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/15.3M [00:00<?, ?B/s]
sub-UCL2_UNIT1.nii.gz: 22%|███▎ | 5.85M/26.7M [00:00<00:01, 17.0MB/s]
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 77.0k/13.6M [00:00<00:25, 565kB/
sub-UCL2_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/832k [00:00<?, ?B/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/15.3M [00:00<00:28, 560kB/
sub-UCL2_UNIT1.nii.gz: 34%|█████▏ | 9.19M/26.7M [00:00<00:00, 22.8MB/s]
sub-UCL2_rec-uncombined2_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 3%| | 383k/13.6M [00:00<00:09, 1.51MB/
sub-UCL2_rec-uncombined1_T2starw.nii.gz: 9%| | 77.0k/832k [00:00<00:01, 552kB/
sub-UCL2_UNIT1.nii.gz: 45%|██████▊ | 12.1M/26.7M [00:00<00:00, 25.3MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 366k/15.3M [00:00<00:10, 1.47MB/
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 9%| | 1.21M/13.6M [00:00<00:03, 4.16MB
sub-UCL2_UNIT1.nii.gz: 56%|████████▍ | 15.1M/26.7M [00:00<00:00, 26.9MB/s]
sub-UCL2_rec-uncombined1_T2starw.nii.gz: 44%|▍| 366k/832k [00:00<00:00, 1.46MB/
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 0.99M/15.3M [00:00<00:04, 3.38MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 16%|▏| 2.23M/13.6M [00:00<00:01, 6.43MB
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 11%| | 1.72M/15.3M [00:00<00:02, 4.86MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 22%|▏| 3.03M/13.6M [00:00<00:01, 7.05MB
sub-UCL2_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/786k [00:00<?, ?B/s]
sub-UCL2_UNIT1.nii.gz: 66%|█████████▉ | 17.7M/26.7M [00:01<00:00, 21.6MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 23%|▏| 3.45M/15.3M [00:00<00:01, 9.26MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 32%|▎| 4.38M/13.6M [00:00<00:01, 9.31MB
sub-UCL2_rec-uncombined2_T2starw.nii.gz: 11%| | 83.6k/786k [00:00<00:01, 616kB/
sub-UCL2_UNIT1.nii.gz: 75%|███████████▏ | 19.9M/26.7M [00:01<00:00, 20.6MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 34%|▎| 5.23M/15.3M [00:00<00:00, 12.3MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 46%|▍| 6.18M/13.6M [00:00<00:00, 12.3MB
sub-UCL2_rec-uncombined3_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined2_T2starw.nii.gz: 28%|▎| 221k/786k [00:00<00:00, 1.01MB/
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 43%|▍| 6.56M/15.3M [00:00<00:00, 12.8MB
sub-UCL2_UNIT1.nii.gz: 82%|████████████▎ | 22.0M/26.7M [00:01<00:00, 19.0MB/s]
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 55%|▌| 7.40M/13.6M [00:00<00:00, 12.4MB
sub-UCL2_rec-uncombined2_T2starw.nii.gz: 99%|▉| 781k/786k [00:00<00:00, 2.86MB/
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 7.89M/15.3M [00:00<00:00, 13.0MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 64%|▋| 8.71M/13.6M [00:00<00:00, 12.8MB
sub-UCL2_UNIT1.nii.gz: 89%|█████████████▍ | 23.9M/26.7M [00:01<00:00, 17.7MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 64%|▋| 9.81M/15.3M [00:00<00:00, 15.1MB
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 79%|▊| 10.8M/13.6M [00:01<00:00, 15.5MB
sub-UCL2_UNIT1.nii.gz: 97%|██████████████▌| 25.9M/26.7M [00:01<00:00, 18.6MB/s]
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 76%|▊| 11.6M/15.3M [00:01<00:00, 15.7MB
sub-UCL2_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/797k [00:00<?, ?B/s]
sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz: 91%|▉| 12.3M/13.6M [00:01<00:00, 15.6MB
sub-UCL2_inv-2_part-mag_MP2RAGE.nii.gz: 90%|▉| 13.8M/15.3M [00:01<00:00, 17.8MB
sub-UCL2_rec-uncombined4_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined3_T2starw.nii.gz: 10%| | 83.5k/797k [00:00<00:01, 594kB/
sub-UCL2_rec-uncombined3_T2starw.nii.gz: 28%|▎| 220k/797k [00:00<00:00, 969kB/s
sub-UCL2_rec-uncombined3_T2starw.nii.gz: 100%|█| 797k/797k [00:00<00:00, 3.00MB/
sub-UCL2_rec-uncombined5_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/751k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined6_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/880k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined4_T2starw.nii.gz: 11%| | 83.6k/751k [00:00<00:01, 612kB/
sub-UCL2_rec-uncombined5_T2starw.nii.gz: 9%| | 83.6k/880k [00:00<00:01, 616kB/
sub-UCL2_rec-uncombined7_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined4_T2starw.nii.gz: 47%|▍| 356k/751k [00:00<00:00, 1.45MB/
sub-UCL2_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/860k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined5_T2starw.nii.gz: 40%|▍| 356k/880k [00:00<00:00, 1.43MB/
sub-UCL2_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/802k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined6_T2starw.nii.gz: 10%| | 83.6k/860k [00:00<00:01, 538kB/
sub-UCL2_rec-uncombined7_T2starw.nii.gz: 10%| | 83.6k/802k [00:00<00:01, 706kB/
sub-UCL2_rec-uncombined6_T2starw.nii.gz: 41%|▍| 356k/860k [00:00<00:00, 1.45MB/
sub-UCL2_rec-uncombined8_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined7_T2starw.nii.gz: 28%|▎| 221k/802k [00:00<00:00, 1.08MB/
sub-UCL2_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/814k [00:00<?, ?B/s]
sub-UCL2_acq-anat_TB1TFL.json: 0%| | 0.00/2.63k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined7_T2starw.nii.gz: 91%|▉| 730k/802k [00:00<00:00, 2.75MB/
sub-UCL2_rec-uncombined8_T2starw.nii.gz: 10%| | 83.5k/814k [00:00<00:01, 612kB/
sub-UCL2_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-UCL2_rec-uncombined8_T2starw.nii.gz: 44%|▍| 356k/814k [00:00<00:00, 1.41MB/
sub-UCL2_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL2_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/797k [00:00<?, ?B/s]
sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/3.92M [00:00<?, ?B/s]
sub-UCL2_acq-anat_TB1TFL.nii.gz: 8%|▌ | 66.6k/797k [00:00<00:01, 555kB/s]
sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 85.2k/3.92M [00:00<00:06, 620kB/
sub-UCL2_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.68M [00:00<?, ?B/s]
sub-UCL2_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.06k [00:00<?, ?B/s]
sub-UCL2_acq-anat_TB1TFL.nii.gz: 40%|██▊ | 322k/797k [00:00<00:00, 1.28MB/s]
sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz: 10%| | 408k/3.92M [00:00<00:02, 1.64MB/
sub-UCL2_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 83.6k/2.68M [00:00<00:04, 61
sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz: 37%|▎| 1.46M/3.92M [00:00<00:00, 5.06MB
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-UCL2_acq-coilQaSagSmall_GFactor.nii.gz: 14%|▏| 373k/2.68M [00:00<00:01, 1.5
sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz: 84%|▊| 3.28M/3.92M [00:00<00:00, 9.75MB
sub-UCL2_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL2_acq-coilQaSagSmall_GFactor.nii.gz: 35%|▎| 951k/2.68M [00:00<00:00, 3.1
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 1%| | 77.0k/11.7M [00:00<00:22, 550kB/s
sub-UCL2_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.4k [00:00<?, ?B/s]
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 3%| | 366k/11.7M [00:00<00:08, 1.48MB/s
sub-UCL2_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL2_acq-famp-0.66_TB1DREAM.nii.gz: 83%|▊| 77.0k/92.4k [00:00<00:00, 571kB/
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 6%| | 774k/11.7M [00:00<00:04, 2.37MB/s
sub-UCL2_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 26%|▎| 3.09M/11.7M [00:00<00:00, 9.81MB/
sub-UCL2_acq-famp_TB1DREAM.json: 0%| | 0.00/2.11k [00:00<?, ?B/s]
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 59%|▌| 6.88M/11.7M [00:00<00:00, 19.6MB/
sub-UCL2_acq-famp-1.5_TB1DREAM.nii.gz: 91%|▉| 83.6k/92.1k [00:00<00:00, 609kB/s
sub-UCL2_acq-coilQaTra_GFactor.nii.gz: 92%|▉| 10.8M/11.7M [00:00<00:00, 25.4MB/
sub-UCL2_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.1k [00:00<?, ?B/s]
sub-UCL2_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/952k [00:00<?, ?B/s]
sub-UCL2_acq-famp_TB1DREAM.nii.gz: 93%|███▋| 85.2k/92.1k [00:00<00:00, 592kB/s]
sub-UCL2_acq-famp_TB1TFL.json: 0%| | 0.00/2.66k [00:00<?, ?B/s]
sub-UCL2_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/952k [00:00<00:01, 626kB/s]
sub-UCL2_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/80.0k [00:00<?, ?B/s]
sub-UCL2_acq-famp_TB1TFL.nii.gz: 37%|██▌ | 356k/952k [00:00<00:00, 1.54MB/s]
sub-UCL2_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-UCL2_acq-refv-0.66_TB1DREAM.nii.gz: 100%|█| 80.0k/80.0k [00:00<00:00, 519kB/
sub-UCL2_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-UCL2_acq-refv_TB1DREAM.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL3_T2starw.json: 0%| | 0.00/2.44k [00:00<?, ?B/s]
sub-UCL2_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/86.6k [00:00<?, ?B/s]
sub-UCL2_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/84.3k [00:00<?, ?B/s]
sub-UCL3_T2starw.nii.gz: 0%| | 0.00/1.06M [00:00<?, ?B/s]
sub-UCL3_UNIT1.nii.gz: 0%| | 0.00/26.3M [00:00<?, ?B/s]
sub-UCL2_acq-refv-1.5_TB1DREAM.nii.gz: 96%|▉| 83.6k/86.6k [00:00<00:00, 681kB/s
sub-UCL2_acq-refv_TB1DREAM.nii.gz: 99%|███▉| 83.6k/84.3k [00:00<00:00, 582kB/s]
sub-UCL3_T2starw.nii.gz: 6%|▊ | 66.6k/1.06M [00:00<00:01, 572kB/s]
sub-UCL3_UNIT1.nii.gz: 0%| | 83.6k/26.3M [00:00<00:44, 613kB/s]
sub-UCL3_T2starw.nii.gz: 23%|███▎ | 254k/1.06M [00:00<00:00, 1.22MB/s]
sub-UCL3_UNIT1.json: 0%| | 0.00/2.12k [00:00<?, ?B/s]
sub-UCL3_UNIT1.nii.gz: 1%|▏ | 238k/26.3M [00:00<00:24, 1.10MB/s]
sub-UCL3_T2starw.nii.gz: 67%|█████████▍ | 730k/1.06M [00:00<00:00, 2.54MB/s]
sub-UCL3_inv-1_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-UCL3_UNIT1.nii.gz: 3%|▍ | 730k/26.3M [00:00<00:10, 2.47MB/s]
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/16.1M [00:00<?, ?B/s]
sub-UCL3_UNIT1.nii.gz: 10%|█▌ | 2.69M/26.3M [00:00<00:03, 8.11MB/s]
sub-UCL3_inv-2_MP2RAGE.json: 0%| | 0.00/2.13k [00:00<?, ?B/s]
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 0%| | 67.5k/16.1M [00:00<00:24, 672kB/
sub-UCL3_UNIT1.nii.gz: 23%|███▍ | 6.04M/26.3M [00:00<00:01, 16.3MB/s]
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 1%| | 169k/16.1M [00:00<00:19, 838kB/s
sub-UCL3_UNIT1.nii.gz: 40%|██████ | 10.6M/26.3M [00:00<00:00, 25.9MB/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 0.00/17.6M [00:00<?, ?B/s]
sub-UCL3_rec-uncombined1_T2starw.nii.gz: 0%| | 0.00/877k [00:00<?, ?B/s]
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 5%| | 747k/16.1M [00:00<00:06, 2.62MB/
sub-UCL3_UNIT1.nii.gz: 54%|████████ | 14.2M/26.3M [00:00<00:00, 29.7MB/s]
sub-UCL3_rec-uncombined2_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 0%| | 77.0k/17.6M [00:00<00:32, 562kB/
sub-UCL3_rec-uncombined1_T2starw.nii.gz: 11%| | 94.0k/877k [00:00<00:01, 689kB/
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 11%| | 1.73M/16.1M [00:00<00:02, 5.35MB
sub-UCL3_UNIT1.nii.gz: 65%|█████████▊ | 17.2M/26.3M [00:00<00:00, 25.9MB/s]
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 26%|▎| 4.10M/16.1M [00:00<00:01, 11.8MB
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 2%| | 383k/17.6M [00:00<00:11, 1.55MB/
sub-UCL3_rec-uncombined1_T2starw.nii.gz: 46%|▍| 400k/877k [00:00<00:00, 1.60MB/
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 37%|▎| 6.02M/16.1M [00:00<00:00, 14.5MB
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 6%| | 995k/17.6M [00:00<00:05, 3.16MB/
sub-UCL3_UNIT1.nii.gz: 75%|███████████▎ | 19.9M/26.3M [00:01<00:00, 23.0MB/s]
sub-UCL3_rec-uncombined1_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 10%| | 1.74M/17.6M [00:00<00:03, 4.79MB
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 46%|▍| 7.46M/16.1M [00:00<00:00, 13.5MB
sub-UCL3_UNIT1.nii.gz: 84%|████████████▋ | 22.2M/26.3M [00:01<00:00, 20.7MB/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 15%|▏| 2.73M/17.6M [00:00<00:02, 6.63MB
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 58%|▌| 9.31M/16.1M [00:00<00:00, 15.2MB
sub-UCL3_UNIT1.nii.gz: 92%|█████████████▊ | 24.3M/26.3M [00:01<00:00, 20.4MB/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 23%|▏| 4.04M/17.6M [00:00<00:01, 8.85MB
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 70%|▋| 11.2M/16.1M [00:00<00:00, 16.6MB
sub-UCL3_rec-uncombined3_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined2_T2starw.nii.gz: 0%| | 0.00/849k [00:00<?, ?B/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 30%|▎| 5.29M/17.6M [00:00<00:01, 10.2MB
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 80%|▊| 12.8M/16.1M [00:01<00:00, 14.9MB
sub-UCL3_rec-uncombined2_T2starw.nii.gz: 10%| | 85.2k/849k [00:00<00:01, 670kB/
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 41%|▍| 7.24M/17.6M [00:00<00:00, 13.3MB
sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz: 96%|▉| 15.5M/16.1M [00:01<00:00, 18.4MB
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 52%|▌| 9.24M/17.6M [00:00<00:00, 15.7MB
sub-UCL3_rec-uncombined2_T2starw.nii.gz: 40%|▍| 340k/849k [00:00<00:00, 1.39MB/
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 76%|▊| 13.3M/17.6M [00:01<00:00, 23.0MB
sub-UCL3_rec-uncombined3_T2starw.nii.gz: 0%| | 0.00/861k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined4_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_inv-2_part-mag_MP2RAGE.nii.gz: 97%|▉| 17.1M/17.6M [00:01<00:00, 27.8MB
sub-UCL3_rec-uncombined3_T2starw.nii.gz: 10%| | 85.2k/861k [00:00<00:01, 654kB/
sub-UCL3_rec-uncombined4_T2starw.nii.gz: 0%| | 0.00/801k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined3_T2starw.nii.gz: 41%|▍| 357k/861k [00:00<00:00, 1.58MB/
sub-UCL3_rec-uncombined5_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined4_T2starw.nii.gz: 12%| | 94.0k/801k [00:00<00:01, 572kB/
sub-UCL3_rec-uncombined5_T2starw.nii.gz: 0%| | 0.00/875k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined6_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined4_T2starw.nii.gz: 50%|▍| 400k/801k [00:00<00:00, 1.66MB/
sub-UCL3_rec-uncombined5_T2starw.nii.gz: 10%| | 83.6k/875k [00:00<00:01, 575kB/
sub-UCL3_rec-uncombined7_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined6_T2starw.nii.gz: 0%| | 0.00/856k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined5_T2starw.nii.gz: 39%|▍| 339k/875k [00:00<00:00, 1.32MB/
sub-UCL3_rec-uncombined7_T2starw.nii.gz: 0%| | 0.00/808k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined6_T2starw.nii.gz: 10%| | 83.6k/856k [00:00<00:01, 726kB/
sub-UCL3_rec-uncombined8_T2starw.json: 0%| | 0.00/2.45k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined7_T2starw.nii.gz: 11%| | 85.2k/808k [00:00<00:01, 646kB/
sub-UCL3_rec-uncombined6_T2starw.nii.gz: 42%|▍| 356k/856k [00:00<00:00, 1.51MB/
sub-UCL3_acq-anat_TB1TFL.json: 0%| | 0.00/2.67k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined7_T2starw.nii.gz: 42%|▍| 340k/808k [00:00<00:00, 1.28MB/
sub-UCL3_acq-anat_TB1TFL.nii.gz: 0%| | 0.00/859k [00:00<?, ?B/s]
sub-UCL3_rec-uncombined8_T2starw.nii.gz: 0%| | 0.00/860k [00:00<?, ?B/s]
sub-UCL3_acq-anat_TB1TFL.nii.gz: 10%|▋ | 83.6k/859k [00:00<00:01, 611kB/s]
sub-UCL3_rec-uncombined8_T2starw.nii.gz: 9%| | 77.0k/860k [00:00<00:01, 659kB/
sub-UCL3_acq-coilQaSagLarge_SNR.json: 0%| | 0.00/2.18k [00:00<?, ?B/s]
sub-UCL3_acq-anat_TB1TFL.nii.gz: 41%|██▉ | 356k/859k [00:00<00:00, 1.42MB/s]
sub-UCL3_rec-uncombined8_T2starw.nii.gz: 43%|▍| 366k/860k [00:00<00:00, 1.57MB/
sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz: 0%| | 0.00/4.17M [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagSmall_GFactor.json: 0%| | 0.00/2.20k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz: 2%| | 83.5k/4.17M [00:00<00:06, 706kB/
sub-UCL3_acq-coilQaSagSmall_GFactor.nii.gz: 0%| | 0.00/2.68M [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz: 8%| | 356k/4.17M [00:00<00:02, 1.50MB/
sub-UCL3_acq-famp-0.66_TB1DREAM.json: 0%| | 0.00/2.15k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaTra_GFactor.json: 0%| | 0.00/2.09k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz: 21%|▏| 901k/4.17M [00:00<00:01, 3.08MB/
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 0%| | 0.00/11.7M [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagSmall_GFactor.nii.gz: 3%| | 85.2k/2.68M [00:00<00:05, 54
sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz: 56%|▌| 2.33M/4.17M [00:00<00:00, 7.16MB
sub-UCL3_acq-coilQaSagSmall_GFactor.nii.gz: 13%|▏| 357k/2.68M [00:00<00:01, 1.4
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 1%| | 83.6k/11.7M [00:00<00:18, 659kB/s
sub-UCL3_acq-coilQaSagSmall_GFactor.nii.gz: 49%|▍| 1.31M/2.68M [00:00<00:00, 4.
sub-UCL3_acq-famp-1.5_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 3%| | 356k/11.7M [00:00<00:08, 1.41MB/s
sub-UCL3_acq-famp-0.66_TB1DREAM.nii.gz: 0%| | 0.00/92.4k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaSagSmall_GFactor.nii.gz: 99%|▉| 2.65M/2.68M [00:00<00:00, 7.
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 12%| | 1.44M/11.7M [00:00<00:02, 4.63MB/
sub-UCL3_acq-famp-0.66_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.4k [00:00<00:00, 614kB/
sub-UCL3_acq-famp-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.8k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 34%|▎| 4.04M/11.7M [00:00<00:00, 11.9MB/
sub-UCL3_acq-famp_TB1DREAM.json: 0%| | 0.00/2.14k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 68%|▋| 8.00M/11.7M [00:00<00:00, 21.2MB/
sub-UCL3_acq-famp-1.5_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.8k [00:00<00:00, 614kB/s
sub-UCL3_acq-famp_TB1DREAM.nii.gz: 0%| | 0.00/92.9k [00:00<?, ?B/s]
sub-UCL3_acq-coilQaTra_GFactor.nii.gz: 96%|▉| 11.2M/11.7M [00:00<00:00, 25.2MB/
sub-UCL3_acq-famp_TB1TFL.json: 0%| | 0.00/2.71k [00:00<?, ?B/s]
sub-UCL3_acq-refv-0.66_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
sub-UCL3_acq-famp_TB1DREAM.nii.gz: 90%|███▌| 83.6k/92.9k [00:00<00:00, 622kB/s]
sub-UCL3_acq-famp_TB1TFL.nii.gz: 0%| | 0.00/949k [00:00<?, ?B/s]
sub-UCL3_acq-refv-1.5_TB1DREAM.json: 0%| | 0.00/2.17k [00:00<?, ?B/s]
sub-UCL3_acq-refv-0.66_TB1DREAM.nii.gz: 0%| | 0.00/87.9k [00:00<?, ?B/s]
sub-UCL3_acq-refv-1.5_TB1DREAM.nii.gz: 0%| | 0.00/92.7k [00:00<?, ?B/s]
sub-UCL3_acq-famp_TB1TFL.nii.gz: 9%|▌ | 83.6k/949k [00:00<00:01, 614kB/s]
sub-UCL3_acq-refv_TB1DREAM.nii.gz: 0%| | 0.00/92.2k [00:00<?, ?B/s]
sub-UCL3_acq-refv-0.66_TB1DREAM.nii.gz: 88%|▉| 77.0k/87.9k [00:00<00:00, 568kB/
sub-UCL3_acq-refv-1.5_TB1DREAM.nii.gz: 90%|▉| 83.6k/92.7k [00:00<00:00, 615kB/s
sub-UCL3_acq-famp_TB1TFL.nii.gz: 37%|██▌ | 356k/949k [00:00<00:00, 1.43MB/s]
sub-UCL3_acq-refv_TB1DREAM.nii.gz: 92%|███▋| 85.2k/92.2k [00:00<00:00, 627kB/s]
sub-UCL3_acq-refv_TB1DREAM.json: 0%| | 0.00/2.16k [00:00<?, ?B/s]
✅ Finished downloading ds005025.
🧠 Please enjoy your brains.
# Define useful variables
path_data = os.path.join(os.getcwd(), "data-human/")
print(f"path_data: {path_data}")
path_labels = os.path.join(path_data, "derivatives", "labels")
path_qc = os.path.join(path_data, "qc")
subjects = [os.path.basename(subject_path) for subject_path in sorted(glob.glob(os.path.join(path_data, "sub-*")))]
print(f"subjects: {subjects}")
# Create output folder
path_results = os.path.join(path_data, "derivatives", "results")
os.makedirs(path_results, exist_ok=True)
path_data: /home/runner/work/coil-qc-code/coil-qc-code/data-human/
subjects: ['sub-CRMBM1', 'sub-CRMBM2', 'sub-CRMBM3', 'sub-MGH1', 'sub-MGH2', 'sub-MGH3', 'sub-MNI1', 'sub-MNI2', 'sub-MNI3', 'sub-MPI1', 'sub-MPI2', 'sub-MPI3', 'sub-MSSM1', 'sub-MSSM2', 'sub-MSSM3', 'sub-NTNU1', 'sub-NTNU2', 'sub-NTNU3', 'sub-UCL1', 'sub-UCL2', 'sub-UCL3']
MP2RAGE segmentation and vertebral labeling#
# Run segmentation on MP2RAGE scan ⏳
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "anat"))
fname_manual_seg = os.path.join(path_labels, subject, "anat", f"{subject}_UNIT1_label-SC_seg.nii.gz")
if os.path.exists(fname_manual_seg):
# Manual segmentation already exists. Copy it to local folder
print(f"{subject}: Manual segmentation found\n")
shutil.copyfile(fname_manual_seg, f"{subject}_UNIT1_seg.nii.gz")
# Generate QC report to make sure the manual segmentation is correct
!sct_qc -i {subject}_UNIT1.nii.gz -s {subject}_UNIT1_seg.nii.gz -p sct_deepseg_sc -qc {path_qc} -qc-subject {subject}
else:
# Manual segmentation does not exist. Run automatic segmentation.
print(f"{subject}: Manual segmentation not found")
!sct_deepseg -i "{subject}_UNIT1.nii.gz" -task seg_sc_contrast_agnostic -thr 0 -qc {path_qc}
sub-CRMBM1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model model_seg_sc_contrast_agnostic_softseg_monai is not installed. Installing it now...
INSTALLING MODEL: model_seg_sc_contrast_agnostic_softseg_monai
Trying URL: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip
Downloading: model_contrast-agnostic_20240930-1002.zip
Status: 0%| | 0.00/431M [00:00<?, ?B/s]
Status: 2%|▊ | 10.5M/431M [00:00<00:04, 95.0MB/s]
Status: 5%|█▌ | 21.0M/431M [00:00<00:04, 99.2MB/s]
Status: 7%|██▎ | 31.5M/431M [00:00<00:04, 99.9MB/s]
Status: 10%|███ | 42.0M/431M [00:00<00:04, 91.1MB/s]
Status: 12%|███▊ | 52.4M/431M [00:00<00:04, 88.2MB/s]
Status: 15%|████▌ | 63.6M/431M [00:00<00:03, 95.4MB/s]
Status: 17%|█████▎ | 73.4M/431M [00:00<00:03, 90.5MB/s]
Status: 19%|█████▉ | 82.6M/431M [00:00<00:04, 82.1MB/s]
Status: 21%|██████▌ | 91.0M/431M [00:01<00:04, 78.6MB/s]
Status: 23%|███████▍ | 100M/431M [00:01<00:03, 82.9MB/s]
Status: 25%|████████ | 109M/431M [00:01<00:04, 76.8MB/s]
Status: 27%|████████▊ | 118M/431M [00:01<00:03, 81.0MB/s]
Status: 29%|█████████▍ | 126M/431M [00:01<00:04, 76.1MB/s]
Status: 32%|██████████▏ | 138M/431M [00:01<00:03, 86.4MB/s]
Status: 35%|███████████ | 149M/431M [00:01<00:03, 92.8MB/s]
Status: 37%|███████████▋ | 158M/431M [00:01<00:02, 91.2MB/s]
Status: 39%|████████████▍ | 168M/431M [00:01<00:02, 89.2MB/s]
Status: 41%|█████████████▏ | 178M/431M [00:02<00:02, 90.6MB/s]
Status: 44%|██████████████ | 189M/431M [00:02<00:02, 88.9MB/s]
Status: 46%|██████████████▋ | 198M/431M [00:02<00:02, 82.6MB/s]
Status: 48%|███████████████▎ | 206M/431M [00:02<00:02, 81.0MB/s]
Status: 50%|███████████████▉ | 214M/431M [00:02<00:02, 76.9MB/s]
Status: 51%|████████████████▍ | 222M/431M [00:02<00:03, 65.8MB/s]
Status: 53%|█████████████████ | 231M/431M [00:02<00:02, 68.9MB/s]
Status: 56%|█████████████████▉ | 241M/431M [00:02<00:02, 74.0MB/s]
Status: 58%|██████████████████▋ | 252M/431M [00:03<00:02, 72.1MB/s]
Status: 61%|███████████████████▍ | 262M/431M [00:03<00:02, 72.2MB/s]
Status: 63%|████████████████████▏ | 273M/431M [00:03<00:02, 77.5MB/s]
Status: 66%|█████████████████████ | 283M/431M [00:03<00:01, 76.2MB/s]
Status: 68%|█████████████████████▊ | 294M/431M [00:03<00:01, 71.1MB/s]
Status: 71%|██████████████████████▌ | 304M/431M [00:03<00:01, 73.1MB/s]
Status: 73%|███████████████████████▎ | 315M/431M [00:03<00:01, 78.6MB/s]
Status: 75%|████████████████████████ | 325M/431M [00:04<00:01, 79.6MB/s]
Status: 78%|████████████████████████▉ | 336M/431M [00:04<00:01, 79.4MB/s]
Status: 80%|█████████████████████████▋ | 347M/431M [00:04<00:00, 87.3MB/s]
Status: 83%|██████████████████████████▍ | 357M/431M [00:04<00:00, 87.3MB/s]
Status: 85%|███████████████████████████▏ | 367M/431M [00:04<00:00, 91.9MB/s]
Status: 88%|████████████████████████████ | 377M/431M [00:04<00:00, 93.0MB/s]
Status: 90%|████████████████████████████▊ | 388M/431M [00:04<00:00, 90.6MB/s]
Status: 92%|█████████████████████████████▌ | 398M/431M [00:04<00:00, 93.9MB/s]
Status: 95%|██████████████████████████████▎ | 409M/431M [00:04<00:00, 87.9MB/s]
Status: 97%|███████████████████████████████ | 419M/431M [00:05<00:00, 89.6MB/s]
Status: 100%|███████████████████████████████▉| 430M/431M [00:05<00:00, 90.4MB/s]
Status: 100%|████████████████████████████████| 431M/431M [00:05<00:00, 83.5MB/s]
Creating temporary folder (/tmp/sct_2025-03-20_01-38-32_install-data_8966trmb)
Unzip data to: /tmp/sct_2025-03-20_01-38-32_install-data_8966trmb
Copying data to: /home/runner/sct_6.5/data/deepseg_models/model_seg_sc_contrast_agnostic_softseg_monai
Removing temporary folders...
Creating temporary folder (/tmp/sct_2025-03-20_01-38-40_sct_deepseg_3k1pvhjz)
Copied sub-CRMBM1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-38-40_sct_deepseg_3k1pvhjz/sub-CRMBM1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 15 seconds
Saving results to: /tmp/sct_2025-03-20_01-38-40_sct_deepseg_3k1pvhjz/sub-CRMBM1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_013857.688035.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-CRMBM2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-39-09_sct_deepseg_22ph0sfn)
Copied sub-CRMBM2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-39-09_sct_deepseg_22ph0sfn/sub-CRMBM2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-39-09_sct_deepseg_22ph0sfn/sub-CRMBM2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_013926.028645.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-CRMBM3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-39-37_sct_deepseg_trtmcynw)
Copied sub-CRMBM3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-39-37_sct_deepseg_trtmcynw/sub-CRMBM3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-39-37_sct_deepseg_trtmcynw/sub-CRMBM3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_013954.099599.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-40-05_sct_deepseg_5hcy5gho)
Copied sub-MGH1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-40-05_sct_deepseg_5hcy5gho/sub-MGH1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 12 seconds
Saving results to: /tmp/sct_2025-03-20_01-40-05_sct_deepseg_5hcy5gho/sub-MGH1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014018.415212.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-40-30_sct_deepseg_7jj16ma8)
Copied sub-MGH2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-40-30_sct_deepseg_7jj16ma8/sub-MGH2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 12 seconds
Saving results to: /tmp/sct_2025-03-20_01-40-30_sct_deepseg_7jj16ma8/sub-MGH2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014043.450851.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-40-56_sct_deepseg_u67dy_0u)
Copied sub-MGH3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-40-56_sct_deepseg_u67dy_0u/sub-MGH3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 12 seconds
Saving results to: /tmp/sct_2025-03-20_01-40-56_sct_deepseg_u67dy_0u/sub-MGH3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014108.425692.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-41-20_sct_deepseg_rq_ojmlh)
Copied sub-MNI1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-41-20_sct_deepseg_rq_ojmlh/sub-MNI1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-41-20_sct_deepseg_rq_ojmlh/sub-MNI1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014137.218530.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-41-49_sct_deepseg_vj35_fuh)
Copied sub-MNI2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-41-49_sct_deepseg_vj35_fuh/sub-MNI2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-41-49_sct_deepseg_vj35_fuh/sub-MNI2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014205.387617.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-42-17_sct_deepseg_35krnwet)
Copied sub-MNI3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-42-17_sct_deepseg_35krnwet/sub-MNI3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-42-17_sct_deepseg_35krnwet/sub-MNI3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014233.366114.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MPI1: Manual segmentation found
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MPI1_UNIT1.nii.gz -s sub-MPI1_UNIT1_seg.nii.gz -p sct_deepseg_sc -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MPI1
--
Resampling image "sub-MPI1_UNIT1.nii.gz" to 0.6x0.6 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Resampling image "sub-MPI1_UNIT1_seg.nii.gz" to 0.6x0.6 mm
Converting image from type 'uint8' to type 'float64' for linear interpolation
Image header specifies datatype 'float64', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Compute center of mass at each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MPI2: Manual segmentation found
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MPI2_UNIT1.nii.gz -s sub-MPI2_UNIT1_seg.nii.gz -p sct_deepseg_sc -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MPI2
--
Resampling image "sub-MPI2_UNIT1.nii.gz" to 0.6x0.6 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Resampling image "sub-MPI2_UNIT1_seg.nii.gz" to 0.6x0.6 mm
Converting image from type 'uint8' to type 'float64' for linear interpolation
Image header specifies datatype 'float64', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Compute center of mass at each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MPI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-42-59_sct_deepseg_csohhcg8)
Copied sub-MPI3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-42-59_sct_deepseg_csohhcg8/sub-MPI3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-42-59_sct_deepseg_csohhcg8/sub-MPI3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014316.083193.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MSSM1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-43-27_sct_deepseg_xgry6ke3)
Copied sub-MSSM1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-43-27_sct_deepseg_xgry6ke3/sub-MSSM1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-43-27_sct_deepseg_xgry6ke3/sub-MSSM1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014344.050707.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MSSM2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-43-55_sct_deepseg_bctejfhf)
Copied sub-MSSM2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-43-55_sct_deepseg_bctejfhf/sub-MSSM2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-43-55_sct_deepseg_bctejfhf/sub-MSSM2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014412.143777.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MSSM3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-44-23_sct_deepseg_m2i2hki2)
Copied sub-MSSM3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-44-23_sct_deepseg_m2i2hki2/sub-MSSM3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-44-23_sct_deepseg_m2i2hki2/sub-MSSM3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014440.108275.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-44-51_sct_deepseg_zmpj84ml)
Copied sub-NTNU1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-44-51_sct_deepseg_zmpj84ml/sub-NTNU1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-44-51_sct_deepseg_zmpj84ml/sub-NTNU1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014508.250306.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-45-20_sct_deepseg_0hqiv5f4)
Copied sub-NTNU2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-45-20_sct_deepseg_0hqiv5f4/sub-NTNU2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-45-20_sct_deepseg_0hqiv5f4/sub-NTNU2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014536.293237.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU3: Manual segmentation found
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-NTNU3_UNIT1.nii.gz -s sub-NTNU3_UNIT1_seg.nii.gz -p sct_deepseg_sc -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-NTNU3
--
Resampling image "sub-NTNU3_UNIT1.nii.gz" to 0.6x0.6 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Resampling image "sub-NTNU3_UNIT1_seg.nii.gz" to 0.6x0.6 mm
Converting image from type 'uint8' to type 'float64' for linear interpolation
Image header specifies datatype 'float64', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Compute center of mass at each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-UCL1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL1_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-45-55_sct_deepseg_mzwheq30)
Copied sub-UCL1_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-45-55_sct_deepseg_mzwheq30/sub-UCL1_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-45-55_sct_deepseg_mzwheq30/sub-UCL1_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014611.677843.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-UCL2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL2_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-46-23_sct_deepseg_7866b740)
Copied sub-UCL2_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-46-23_sct_deepseg_7866b740/sub-UCL2_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-46-23_sct_deepseg_7866b740/sub-UCL2_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014639.620045.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-UCL3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL3_UNIT1.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-46-51_sct_deepseg_mqsnjfrm)
Copied sub-UCL3_UNIT1.nii.gz to /tmp/sct_2025-03-20_01-46-51_sct_deepseg_mqsnjfrm/sub-UCL3_UNIT1.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 14 seconds
Saving results to: /tmp/sct_2025-03-20_01-46-51_sct_deepseg_mqsnjfrm/sub-UCL3_UNIT1_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_014707.524177.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
# Label vertebrae
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "anat"))
fname_manual_labels = os.path.join(path_labels, subject, "anat", f"{subject}_UNIT1_label-disc.nii.gz")
if os.path.exists(fname_manual_labels):
# Use manual disc labels to generate labeled segmentation.
print(f"{subject}: Manual labels found\n")
!sct_label_utils -i {subject}_UNIT1_seg.nii.gz -disc {fname_manual_labels} -o {subject}_UNIT1_seg_labeled.nii.gz
# Generate QC report to assess labeled segmentation
!sct_qc -i {subject}_UNIT1.nii.gz -s {subject}_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc {path_qc} -qc-subject {subject}
else:
# Manual labels do not exist. They should!
print(f"{subject}: Manual labels not found.")
sub-CRMBM1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-CRMBM1/anat/sub-CRMBM1_UNIT1_label-disc.nii.gz -o sub-CRMBM1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-CRMBM1_UNIT1.nii.gz -s sub-CRMBM1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-CRMBM1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-CRMBM2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-CRMBM2/anat/sub-CRMBM2_UNIT1_label-disc.nii.gz -o sub-CRMBM2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-CRMBM2_UNIT1.nii.gz -s sub-CRMBM2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-CRMBM2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-CRMBM3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-CRMBM3/anat/sub-CRMBM3_UNIT1_label-disc.nii.gz -o sub-CRMBM3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-CRMBM3_UNIT1.nii.gz -s sub-CRMBM3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-CRMBM3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MGH1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MGH1/anat/sub-MGH1_UNIT1_label-disc.nii.gz -o sub-MGH1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MGH1_UNIT1.nii.gz -s sub-MGH1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MGH1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MGH2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MGH2/anat/sub-MGH2_UNIT1_label-disc.nii.gz -o sub-MGH2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MGH2_UNIT1.nii.gz -s sub-MGH2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MGH2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MGH3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MGH3/anat/sub-MGH3_UNIT1_label-disc.nii.gz -o sub-MGH3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MGH3_UNIT1.nii.gz -s sub-MGH3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MGH3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MNI1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MNI1/anat/sub-MNI1_UNIT1_label-disc.nii.gz -o sub-MNI1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MNI1_UNIT1.nii.gz -s sub-MNI1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MNI1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MNI2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MNI2/anat/sub-MNI2_UNIT1_label-disc.nii.gz -o sub-MNI2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MNI2_UNIT1.nii.gz -s sub-MNI2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MNI2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MNI3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MNI3/anat/sub-MNI3_UNIT1_label-disc.nii.gz -o sub-MNI3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MNI3_UNIT1.nii.gz -s sub-MNI3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MNI3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MPI1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MPI1/anat/sub-MPI1_UNIT1_label-disc.nii.gz -o sub-MPI1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MPI1_UNIT1.nii.gz -s sub-MPI1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MPI1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MPI2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MPI2/anat/sub-MPI2_UNIT1_label-disc.nii.gz -o sub-MPI2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MPI2_UNIT1.nii.gz -s sub-MPI2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MPI2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MPI3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MPI3/anat/sub-MPI3_UNIT1_label-disc.nii.gz -o sub-MPI3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MPI3_UNIT1.nii.gz -s sub-MPI3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MPI3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MSSM1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MSSM1/anat/sub-MSSM1_UNIT1_label-disc.nii.gz -o sub-MSSM1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MSSM1_UNIT1.nii.gz -s sub-MSSM1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MSSM1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MSSM2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MSSM2/anat/sub-MSSM2_UNIT1_label-disc.nii.gz -o sub-MSSM2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MSSM2_UNIT1.nii.gz -s sub-MSSM2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MSSM2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-MSSM3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-MSSM3/anat/sub-MSSM3_UNIT1_label-disc.nii.gz -o sub-MSSM3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-MSSM3_UNIT1.nii.gz -s sub-MSSM3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-MSSM3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-NTNU1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-NTNU1/anat/sub-NTNU1_UNIT1_label-disc.nii.gz -o sub-NTNU1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-NTNU1_UNIT1.nii.gz -s sub-NTNU1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-NTNU1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-NTNU2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-NTNU2/anat/sub-NTNU2_UNIT1_label-disc.nii.gz -o sub-NTNU2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-NTNU2_UNIT1.nii.gz -s sub-NTNU2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-NTNU2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-NTNU3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-NTNU3/anat/sub-NTNU3_UNIT1_label-disc.nii.gz -o sub-NTNU3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-NTNU3_UNIT1.nii.gz -s sub-NTNU3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-NTNU3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-UCL1: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL1_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-UCL1/anat/sub-UCL1_UNIT1_label-disc.nii.gz -o sub-UCL1_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-UCL1_UNIT1.nii.gz -s sub-UCL1_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-UCL1
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-UCL2: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL2_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-UCL2/anat/sub-UCL2_UNIT1_label-disc.nii.gz -o sub-UCL2_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-UCL2_UNIT1.nii.gz -s sub-UCL2_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-UCL2
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-UCL3: Manual labels found
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL3_UNIT1_seg.nii.gz -disc /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/labels/sub-UCL3/anat/sub-UCL3_UNIT1_label-disc.nii.gz -o sub-UCL3_UNIT1_seg_labeled.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-UCL3_UNIT1.nii.gz -s sub-UCL3_UNIT1_seg_labeled.nii.gz -p sct_label_vertebrae -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-UCL3
--
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
# Crop MP2RAGE for faster processing and better registration results
dilation_kernel="20x20x0"
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "anat"))
!sct_crop_image -i {subject}_inv-1_part-mag_MP2RAGE.nii.gz -m {subject}_UNIT1_seg.nii.gz -dilate {dilation_kernel} -o {subject}_inv-1_part-mag_MP2RAGE_crop.nii.gz
!sct_crop_image -i {subject}_UNIT1.nii.gz -m {subject}_UNIT1_seg.nii.gz -dilate {dilation_kernel} -o {subject}_UNIT1_crop.nii.gz
!sct_crop_image -i {subject}_UNIT1_seg.nii.gz -m {subject}_UNIT1_seg.nii.gz -dilate {dilation_kernel} -o {subject}_UNIT1_seg_crop.nii.gz
!sct_crop_image -i {subject}_UNIT1_seg_labeled.nii.gz -m {subject}_UNIT1_seg.nii.gz -dilate {dilation_kernel} -o {subject}_UNIT1_seg_labeled_crop.nii.gz
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-CRMBM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[92, 157], y=[24, 124], z=[54, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_UNIT1.nii.gz -m sub-CRMBM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM1_UNIT1_crop.nii.gz
--
Bounding box: x=[92, 157], y=[24, 124], z=[54, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_UNIT1_seg.nii.gz -m sub-CRMBM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[92, 157], y=[24, 124], z=[54, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_UNIT1_seg_labeled.nii.gz -m sub-CRMBM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[92, 157], y=[24, 124], z=[54, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-CRMBM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[88, 153], y=[33, 123], z=[63, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_UNIT1.nii.gz -m sub-CRMBM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM2_UNIT1_crop.nii.gz
--
Bounding box: x=[88, 153], y=[33, 123], z=[63, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_UNIT1_seg.nii.gz -m sub-CRMBM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[88, 153], y=[33, 123], z=[63, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_UNIT1_seg_labeled.nii.gz -m sub-CRMBM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[88, 153], y=[33, 123], z=[63, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-CRMBM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[84, 153], y=[14, 135], z=[0, 337]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_UNIT1.nii.gz -m sub-CRMBM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM3_UNIT1_crop.nii.gz
--
Bounding box: x=[84, 153], y=[14, 135], z=[0, 337]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_UNIT1_seg.nii.gz -m sub-CRMBM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[84, 153], y=[14, 135], z=[0, 337]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_UNIT1_seg_labeled.nii.gz -m sub-CRMBM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[84, 153], y=[14, 135], z=[0, 337]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MGH1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[63, 125], y=[42, 160], z=[30, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_UNIT1.nii.gz -m sub-MGH1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH1_UNIT1_crop.nii.gz
--
Bounding box: x=[63, 125], y=[42, 160], z=[30, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_UNIT1_seg.nii.gz -m sub-MGH1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[63, 125], y=[42, 160], z=[30, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_UNIT1_seg_labeled.nii.gz -m sub-MGH1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[63, 125], y=[42, 160], z=[30, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MGH2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[65, 124], y=[34, 151], z=[20, 243]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_UNIT1.nii.gz -m sub-MGH2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH2_UNIT1_crop.nii.gz
--
Bounding box: x=[65, 124], y=[34, 151], z=[20, 243]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_UNIT1_seg.nii.gz -m sub-MGH2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[65, 124], y=[34, 151], z=[20, 243]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_UNIT1_seg_labeled.nii.gz -m sub-MGH2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[65, 124], y=[34, 151], z=[20, 243]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MGH3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[61, 124], y=[38, 165], z=[18, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_UNIT1.nii.gz -m sub-MGH3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH3_UNIT1_crop.nii.gz
--
Bounding box: x=[61, 124], y=[38, 165], z=[18, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_UNIT1_seg.nii.gz -m sub-MGH3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[61, 124], y=[38, 165], z=[18, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_UNIT1_seg_labeled.nii.gz -m sub-MGH3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MGH3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[61, 124], y=[38, 165], z=[18, 256]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MNI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[90, 155], y=[49, 126], z=[0, 367]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_UNIT1.nii.gz -m sub-MNI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI1_UNIT1_crop.nii.gz
--
Bounding box: x=[90, 155], y=[49, 126], z=[0, 367]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_UNIT1_seg.nii.gz -m sub-MNI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[90, 155], y=[49, 126], z=[0, 367]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_UNIT1_seg_labeled.nii.gz -m sub-MNI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[90, 155], y=[49, 126], z=[0, 367]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MNI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[91, 154], y=[39, 114], z=[0, 361]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_UNIT1.nii.gz -m sub-MNI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI2_UNIT1_crop.nii.gz
--
Bounding box: x=[91, 154], y=[39, 114], z=[0, 361]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_UNIT1_seg.nii.gz -m sub-MNI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[91, 154], y=[39, 114], z=[0, 361]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_UNIT1_seg_labeled.nii.gz -m sub-MNI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[91, 154], y=[39, 114], z=[0, 361]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MNI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[81, 150], y=[60, 162], z=[19, 279]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_UNIT1.nii.gz -m sub-MNI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI3_UNIT1_crop.nii.gz
--
Bounding box: x=[81, 150], y=[60, 162], z=[19, 279]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_UNIT1_seg.nii.gz -m sub-MNI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[81, 150], y=[60, 162], z=[19, 279]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_UNIT1_seg_labeled.nii.gz -m sub-MNI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MNI3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[81, 150], y=[60, 162], z=[19, 279]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MPI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[96, 164], y=[29, 118], z=[9, 275]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_UNIT1.nii.gz -m sub-MPI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI1_UNIT1_crop.nii.gz
--
Bounding box: x=[96, 164], y=[29, 118], z=[9, 275]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_UNIT1_seg.nii.gz -m sub-MPI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[96, 164], y=[29, 118], z=[9, 275]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_UNIT1_seg_labeled.nii.gz -m sub-MPI1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[96, 164], y=[29, 118], z=[9, 275]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MPI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[87, 148], y=[31, 120], z=[0, 276]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_UNIT1.nii.gz -m sub-MPI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI2_UNIT1_crop.nii.gz
--
Bounding box: x=[87, 148], y=[31, 120], z=[0, 276]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_UNIT1_seg.nii.gz -m sub-MPI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[87, 148], y=[31, 120], z=[0, 276]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_UNIT1_seg_labeled.nii.gz -m sub-MPI2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[87, 148], y=[31, 120], z=[0, 276]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MPI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[93, 161], y=[14, 131], z=[7, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_UNIT1.nii.gz -m sub-MPI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI3_UNIT1_crop.nii.gz
--
Bounding box: x=[93, 161], y=[14, 131], z=[7, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_UNIT1_seg.nii.gz -m sub-MPI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[93, 161], y=[14, 131], z=[7, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_UNIT1_seg_labeled.nii.gz -m sub-MPI3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MPI3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[93, 161], y=[14, 131], z=[7, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MSSM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[81, 149], y=[50, 122], z=[16, 254]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_UNIT1.nii.gz -m sub-MSSM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM1_UNIT1_crop.nii.gz
--
Bounding box: x=[81, 149], y=[50, 122], z=[16, 254]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_UNIT1_seg.nii.gz -m sub-MSSM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[81, 149], y=[50, 122], z=[16, 254]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_UNIT1_seg_labeled.nii.gz -m sub-MSSM1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[81, 149], y=[50, 122], z=[16, 254]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MSSM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[88, 150], y=[50, 124], z=[40, 268]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_UNIT1.nii.gz -m sub-MSSM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM2_UNIT1_crop.nii.gz
--
Bounding box: x=[88, 150], y=[50, 124], z=[40, 268]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_UNIT1_seg.nii.gz -m sub-MSSM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[88, 150], y=[50, 124], z=[40, 268]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_UNIT1_seg_labeled.nii.gz -m sub-MSSM2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[88, 150], y=[50, 124], z=[40, 268]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-MSSM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[87, 152], y=[32, 136], z=[0, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_UNIT1.nii.gz -m sub-MSSM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM3_UNIT1_crop.nii.gz
--
Bounding box: x=[87, 152], y=[32, 136], z=[0, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_UNIT1_seg.nii.gz -m sub-MSSM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[87, 152], y=[32, 136], z=[0, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_UNIT1_seg_labeled.nii.gz -m sub-MSSM3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[87, 152], y=[32, 136], z=[0, 289]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-NTNU1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[90, 155], y=[61, 126], z=[20, 316]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_UNIT1.nii.gz -m sub-NTNU1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU1_UNIT1_crop.nii.gz
--
Bounding box: x=[90, 155], y=[61, 126], z=[20, 316]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_UNIT1_seg.nii.gz -m sub-NTNU1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[90, 155], y=[61, 126], z=[20, 316]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_UNIT1_seg_labeled.nii.gz -m sub-NTNU1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[90, 155], y=[61, 126], z=[20, 316]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-NTNU2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[82, 151], y=[49, 120], z=[60, 294]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_UNIT1.nii.gz -m sub-NTNU2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU2_UNIT1_crop.nii.gz
--
Bounding box: x=[82, 151], y=[49, 120], z=[60, 294]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_UNIT1_seg.nii.gz -m sub-NTNU2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[82, 151], y=[49, 120], z=[60, 294]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_UNIT1_seg_labeled.nii.gz -m sub-NTNU2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[82, 151], y=[49, 120], z=[60, 294]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-NTNU3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[94, 154], y=[23, 126], z=[8, 330]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_UNIT1.nii.gz -m sub-NTNU3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU3_UNIT1_crop.nii.gz
--
Bounding box: x=[94, 154], y=[23, 126], z=[8, 330]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_UNIT1_seg.nii.gz -m sub-NTNU3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[94, 154], y=[23, 126], z=[8, 330]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_UNIT1_seg_labeled.nii.gz -m sub-NTNU3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[94, 154], y=[23, 126], z=[8, 330]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_inv-1_part-mag_MP2RAGE.nii.gz -m sub-UCL1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[103, 170], y=[18, 123], z=[35, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_UNIT1.nii.gz -m sub-UCL1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL1_UNIT1_crop.nii.gz
--
Bounding box: x=[103, 170], y=[18, 123], z=[35, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_UNIT1_seg.nii.gz -m sub-UCL1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL1_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[103, 170], y=[18, 123], z=[35, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_UNIT1_seg_labeled.nii.gz -m sub-UCL1_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL1_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[103, 170], y=[18, 123], z=[35, 368]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_inv-1_part-mag_MP2RAGE.nii.gz -m sub-UCL2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[77, 152], y=[25, 135], z=[72, 359]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_UNIT1.nii.gz -m sub-UCL2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL2_UNIT1_crop.nii.gz
--
Bounding box: x=[77, 152], y=[25, 135], z=[72, 359]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_UNIT1_seg.nii.gz -m sub-UCL2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL2_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[77, 152], y=[25, 135], z=[72, 359]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_UNIT1_seg_labeled.nii.gz -m sub-UCL2_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL2_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[77, 152], y=[25, 135], z=[72, 359]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_inv-1_part-mag_MP2RAGE.nii.gz -m sub-UCL3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz
--
Bounding box: x=[91, 160], y=[12, 127], z=[62, 362]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_UNIT1.nii.gz -m sub-UCL3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL3_UNIT1_crop.nii.gz
--
Bounding box: x=[91, 160], y=[12, 127], z=[62, 362]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_UNIT1_seg.nii.gz -m sub-UCL3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL3_UNIT1_seg_crop.nii.gz
--
Bounding box: x=[91, 160], y=[12, 127], z=[62, 362]
Cropping the image...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_UNIT1_seg_labeled.nii.gz -m sub-UCL3_UNIT1_seg.nii.gz -dilate 20x20x0 -o sub-UCL3_UNIT1_seg_labeled_crop.nii.gz
--
Bounding box: x=[91, 160], y=[12, 127], z=[62, 362]
Cropping the image...
Register MP2RAGE to B1+ and SNR data#
Segment spinal cord on TFL#
# Segment spinal cord on TFL data (B1+ mapping) ⏳
# Only do this for subjects without MSSM in their name (https://github.com/spinal-cord-7t/coil-qc-code/issues/153)
subjects_without_MSSM = [subject for subject in subjects if "MSSM" not in subject]
for subject in subjects_without_MSSM:
os.chdir(os.path.join(path_data, subject, "fmap"))
fname_manual_seg = os.path.join(path_labels, subject, "fmap", f"{subject}_acq-anat_TB1TFL_label-SC_seg.nii.gz")
if os.path.exists(fname_manual_seg):
# Manual segmentation already exists. Copy it to local folder
print(f"{subject}: Manual segmentation found\n")
shutil.copyfile(fname_manual_seg, f"{subject}_acq-anat_TB1TFL_seg.nii.gz")
# Generate QC report to make sure the manual segmentation is correct
if subject in ['sub-MSSM1', 'sub-MSSM2', 'sub-MSSM3']:
!sct_qc -i "{subject}_acq-famp_TB1TFL.nii.gz" -s "{subject}_acq-anat_TB1TFL_seg.nii.gz" -p sct_deepseg_sc -qc {path_qc} -qc-subject {subject}
else:
!sct_qc -i "{subject}_acq-anat_TB1TFL.nii.gz" -s "{subject}_acq-anat_TB1TFL_seg.nii.gz" -p sct_deepseg_sc -qc {path_qc} -qc-subject {subject}
else:
# Manual segmentation does not exist. Run automatic segmentation.
print(f"{subject}: Manual segmentation not found")
!sct_deepseg -i "{subject}_acq-anat_TB1TFL.nii.gz" -task seg_sc_contrast_agnostic -thr 0 -qc {path_qc}
sub-CRMBM1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-49-50_sct_deepseg_u43djoy8)
Copied sub-CRMBM1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-49-50_sct_deepseg_u43djoy8/sub-CRMBM1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 24 seconds
Saving results to: /tmp/sct_2025-03-20_01-49-50_sct_deepseg_u43djoy8/sub-CRMBM1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015016.367036.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-CRMBM2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-50-24_sct_deepseg_7z4kt2lt)
Copied sub-CRMBM2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-50-24_sct_deepseg_7z4kt2lt/sub-CRMBM2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 24 seconds
Saving results to: /tmp/sct_2025-03-20_01-50-24_sct_deepseg_7z4kt2lt/sub-CRMBM2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015049.815987.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-CRMBM3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM3_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-50-58_sct_deepseg_9zch60k6)
Copied sub-CRMBM3_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-50-58_sct_deepseg_9zch60k6/sub-CRMBM3_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-50-58_sct_deepseg_9zch60k6/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015123.008864.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-51-31_sct_deepseg_tmt702xn)
Copied sub-MGH1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-51-31_sct_deepseg_tmt702xn/sub-MGH1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-51-31_sct_deepseg_tmt702xn/sub-MGH1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015156.398700.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-52-04_sct_deepseg_r_p6wy33)
Copied sub-MGH2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-52-04_sct_deepseg_r_p6wy33/sub-MGH2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-52-04_sct_deepseg_r_p6wy33/sub-MGH2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015229.639762.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MGH3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH3_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-52-38_sct_deepseg_v_uxo92q)
Copied sub-MGH3_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-52-38_sct_deepseg_v_uxo92q/sub-MGH3_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-52-38_sct_deepseg_v_uxo92q/sub-MGH3_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015303.008781.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-53-11_sct_deepseg_xnmx9khe)
Copied sub-MNI1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-53-11_sct_deepseg_xnmx9khe/sub-MNI1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-53-11_sct_deepseg_xnmx9khe/sub-MNI1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015336.173016.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-53-44_sct_deepseg_ldg3mqi9)
Copied sub-MNI2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-53-44_sct_deepseg_ldg3mqi9/sub-MNI2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-53-44_sct_deepseg_ldg3mqi9/sub-MNI2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015409.295345.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MNI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI3_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-54-17_sct_deepseg_lb2fm1jr)
Copied sub-MNI3_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-54-17_sct_deepseg_lb2fm1jr/sub-MNI3_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-54-17_sct_deepseg_lb2fm1jr/sub-MNI3_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015442.458109.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MPI1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-54-50_sct_deepseg_xj2_r_sa)
Copied sub-MPI1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-54-50_sct_deepseg_xj2_r_sa/sub-MPI1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-54-50_sct_deepseg_xj2_r_sa/sub-MPI1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015515.764258.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MPI2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-55-24_sct_deepseg_icp_1nao)
Copied sub-MPI2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-55-24_sct_deepseg_icp_1nao/sub-MPI2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-55-24_sct_deepseg_icp_1nao/sub-MPI2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015548.921869.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-MPI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI3_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-55-57_sct_deepseg_p6r3qd7g)
Copied sub-MPI3_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-55-57_sct_deepseg_p6r3qd7g/sub-MPI3_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-55-57_sct_deepseg_p6r3qd7g/sub-MPI3_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015622.250217.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-56-30_sct_deepseg_rmyaaea3)
Copied sub-NTNU1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-56-30_sct_deepseg_rmyaaea3/sub-NTNU1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-56-30_sct_deepseg_rmyaaea3/sub-NTNU1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015655.502840.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-57-03_sct_deepseg_72s_439q)
Copied sub-NTNU2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-57-03_sct_deepseg_72s_439q/sub-NTNU2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-57-03_sct_deepseg_72s_439q/sub-NTNU2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015728.826621.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-NTNU3: Manual segmentation found
--
Spinal Cord Toolbox (6.5)
sct_qc -i sub-NTNU3_acq-anat_TB1TFL.nii.gz -s sub-NTNU3_acq-anat_TB1TFL_seg.nii.gz -p sct_deepseg_sc -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc -qc-subject sub-NTNU3
--
Resampling image "sub-NTNU3_acq-anat_TB1TFL.nii.gz" to 0.6x0.6 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Resampling image "sub-NTNU3_acq-anat_TB1TFL_seg.nii.gz" to 0.6x0.6 mm
Converting image from type 'uint8' to type 'float64' for linear interpolation
Image header specifies datatype 'float64', but array is of type 'int64'. Header metadata will be overwritten to use 'int64'.
Compute center of mass at each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
*** Generating Quality Control (QC) html report ***
Traceback (most recent call last):
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 125, in <module>
main(sys.argv[1:])
File "/home/runner/sct_6.5/spinalcordtoolbox/scripts/sct_qc.py", line 108, in main
generate_qc(fname_in1=arguments.i,
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 632, in generate_qc
qc_image._make_QC_image_for_3d_volumes(img, mask, plane, imgs_to_generate)
File "/home/runner/sct_6.5/spinalcordtoolbox/reports/qc.py", line 228, in _make_QC_image_for_3d_volumes
fig = mpl_plt.figure()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 1027, in figure
manager = new_figure_manager(
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 549, in new_figure_manager
_warn_if_gui_out_of_main_thread()
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 526, in _warn_if_gui_out_of_main_thread
canvas_class = cast(type[FigureCanvasBase], _get_backend_mod().FigureCanvas)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 358, in _get_backend_mod
switch_backend(rcParams._get("backend"))
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/pyplot.py", line 415, in switch_backend
module = backend_registry.load_backend_module(newbackend)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib/backends/registry.py", line 323, in load_backend_module
return importlib.import_module(module_name)
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 972, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 986, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 680, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 850, in exec_module
File "<frozen importlib._bootstrap>", line 228, in _call_with_frames_removed
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/__init__.py", line 1, in <module>
from . import backend_inline, config # noqa
File "/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/matplotlib_inline/backend_inline.py", line 13, in <module>
from IPython.core.interactiveshell import InteractiveShell
ModuleNotFoundError: No module named 'IPython'
sub-UCL1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL1_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-57-40_sct_deepseg_mzla8lio)
Copied sub-UCL1_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-57-40_sct_deepseg_mzla8lio/sub-UCL1_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-57-40_sct_deepseg_mzla8lio/sub-UCL1_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015805.442144.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-UCL2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL2_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-58-13_sct_deepseg_e8km59oo)
Copied sub-UCL2_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-58-13_sct_deepseg_e8km59oo/sub-UCL2_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-58-13_sct_deepseg_e8km59oo/sub-UCL2_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015838.736284.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
sub-UCL3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL3_acq-anat_TB1TFL.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_01-58-47_sct_deepseg_ue87p1so)
Copied sub-UCL3_acq-anat_TB1TFL.nii.gz to /tmp/sct_2025-03-20_01-58-47_sct_deepseg_ue87p1so/sub-UCL3_acq-anat_TB1TFL.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_01-58-47_sct_deepseg_ue87p1so/sub-UCL3_acq-anat_TB1TFL_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015912.139220.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
Register TFL to MP2RAGE#
# Register TFL data (B1+ mapping) to the MP2RAGE scan ⏳
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
if subject in ['sub-MSSM1', 'sub-MSSM2', 'sub-MSSM3']:
# https://github.com/spinal-cord-7t/coil-qc-code/issues/43
# https://github.com/spinal-cord-7t/coil-qc-code/issues/153
!sct_register_multimodal -i {subject}_acq-famp_TB1TFL.nii.gz -d ../anat/{subject}_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/{subject}_UNIT1_seg_crop.nii.gz -identity 1 -qc "{path_qc}"
else:
!sct_register_multimodal -i {subject}_acq-anat_TB1TFL.nii.gz -iseg {subject}_acq-anat_TB1TFL_seg.nii.gz -d ../anat/{subject}_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/{subject}_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc "{path_qc}"
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM1_acq-anat_TB1TFL.nii.gz -iseg sub-CRMBM1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-CRMBM1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM1_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 100, 314)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_01-59-19_register-slicewise_ywrkrpq_)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 100 x 314
voxel size: 0.7065217mm x 0.6999998mm x 314mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/314 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/314 [00:00<01:08, 4.54iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1446: RuntimeWarning: invalid value encountered in divide
coordsrc /= coordsrc.std()
Slice #8 is empty. It will be ignored.
Estimate cord angle for each slice: 39%|████████▌ | 123/314 [00:00<00:00, 482.45iter/s]
Estimate cord angle for each slice: 79%|█████████████████▎ | 247/314 [00:00<00:00, 754.31iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #311 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #312 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #313 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 314/314 [00:00<00:00, 660.42iter/s]
Build 3D deformation field: 0%| | 0/302 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 13/302 [00:00<00:02, 122.74iter/s]
Build 3D deformation field: 9%|██▊ | 27/302 [00:00<00:02, 127.98iter/s]
Build 3D deformation field: 14%|████▏ | 41/302 [00:00<00:02, 129.68iter/s]
Build 3D deformation field: 18%|█████▋ | 55/302 [00:00<00:01, 130.48iter/s]
Build 3D deformation field: 23%|███████ | 69/302 [00:00<00:01, 132.18iter/s]
Build 3D deformation field: 27%|████████▌ | 83/302 [00:00<00:01, 132.29iter/s]
Build 3D deformation field: 32%|█████████▉ | 97/302 [00:00<00:01, 132.40iter/s]
Build 3D deformation field: 37%|███████████ | 111/302 [00:00<00:01, 133.23iter/s]
Build 3D deformation field: 41%|████████████▍ | 125/302 [00:00<00:01, 133.23iter/s]
Build 3D deformation field: 46%|█████████████▊ | 139/302 [00:01<00:01, 134.08iter/s]
Build 3D deformation field: 51%|███████████████▏ | 153/302 [00:01<00:01, 135.54iter/s]
Build 3D deformation field: 55%|████████████████▌ | 167/302 [00:01<00:00, 136.12iter/s]
Build 3D deformation field: 60%|█████████████████▉ | 181/302 [00:01<00:00, 136.90iter/s]
Build 3D deformation field: 65%|███████████████████▎ | 195/302 [00:01<00:00, 135.50iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 209/302 [00:01<00:00, 134.92iter/s]
Build 3D deformation field: 74%|██████████████████████▏ | 223/302 [00:01<00:00, 134.70iter/s]
Build 3D deformation field: 78%|███████████████████████▌ | 237/302 [00:01<00:00, 133.86iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 251/302 [00:01<00:00, 133.83iter/s]
Build 3D deformation field: 88%|██████████████████████████▎ | 265/302 [00:01<00:00, 133.48iter/s]
Build 3D deformation field: 92%|███████████████████████████▋ | 279/302 [00:02<00:00, 133.53iter/s]
Build 3D deformation field: 97%|█████████████████████████████ | 293/302 [00:02<00:00, 133.42iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 302/302 [00:02<00:00, 133.41iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
rm -rf /tmp/sct_2025-03-20_01-59-19_register-slicewise_ywrkrpq_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh/warp_src2dest.nii.gz warp_sub-CRMBM1_acq-anat_TB1TFL2sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM1_acq-anat_TB1TFL2sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh/warp_dest2src.nii.gz warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_01-59-16_register-wrapper_m92hffeh
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015925.670363.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM2_acq-anat_TB1TFL.nii.gz -iseg sub-CRMBM2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-CRMBM2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM2_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 90, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_01-59-30_register-slicewise_53hdfja6)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 90 x 226
voxel size: 0.7065217mm x 0.6999998mm x 226mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/226 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/226 [00:00<00:48, 4.64iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
Estimate cord angle for each slice: 55%|████████████▏ | 125/226 [00:00<00:00, 495.80iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #224 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #225 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 226/226 [00:00<00:00, 568.49iter/s]
Build 3D deformation field: 0%| | 0/220 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▉ | 14/220 [00:00<00:01, 138.85iter/s]
Build 3D deformation field: 13%|████ | 29/220 [00:00<00:01, 143.27iter/s]
Build 3D deformation field: 20%|██████▏ | 44/220 [00:00<00:01, 145.00iter/s]
Build 3D deformation field: 27%|████████▍ | 60/220 [00:00<00:01, 148.38iter/s]
Build 3D deformation field: 35%|██████████▋ | 76/220 [00:00<00:00, 149.50iter/s]
Build 3D deformation field: 41%|████████████▊ | 91/220 [00:00<00:00, 149.43iter/s]
Build 3D deformation field: 49%|██████████████▌ | 107/220 [00:00<00:00, 149.63iter/s]
Build 3D deformation field: 55%|████████████████▋ | 122/220 [00:00<00:00, 148.83iter/s]
Build 3D deformation field: 62%|██████████████████▋ | 137/220 [00:00<00:00, 148.24iter/s]
Build 3D deformation field: 69%|████████████████████▋ | 152/220 [00:01<00:00, 147.95iter/s]
Build 3D deformation field: 76%|██████████████████████▊ | 167/220 [00:01<00:00, 147.91iter/s]
Build 3D deformation field: 83%|████████████████████████▊ | 182/220 [00:01<00:00, 147.93iter/s]
Build 3D deformation field: 90%|███████████████████████████ | 198/220 [00:01<00:00, 148.95iter/s]
Build 3D deformation field: 97%|█████████████████████████████ | 213/220 [00:01<00:00, 148.14iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 220/220 [00:01<00:00, 147.93iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
rm -rf /tmp/sct_2025-03-20_01-59-30_register-slicewise_53hdfja6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_/warp_src2dest.nii.gz warp_sub-CRMBM2_acq-anat_TB1TFL2sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM2_acq-anat_TB1TFL2sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_/warp_dest2src.nii.gz warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_01-59-28_register-wrapper_h505log_
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015934.783788.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM3_acq-anat_TB1TFL.nii.gz -iseg sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-CRMBM3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM3_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 121, 337)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_01-59-40_register-slicewise_0ktrmpdh)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 121 x 337
voxel size: 0.7065217mm x 0.6999998mm x 337mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/337 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/337 [00:00<01:14, 4.54iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #37 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #38 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #39 is empty. It will be ignored.
Estimate cord angle for each slice: 33%|███████▏ | 110/337 [00:00<00:00, 430.63iter/s]
Estimate cord angle for each slice: 68%|███████████████ | 230/337 [00:00<00:00, 705.72iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #324 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #325 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #326 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #327 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #328 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #329 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #330 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #331 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #332 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #333 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #334 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #335 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #336 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 337/337 [00:00<00:00, 665.39iter/s]
Build 3D deformation field: 0%| | 0/284 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▏ | 10/284 [00:00<00:02, 97.69iter/s]
Build 3D deformation field: 7%|██▎ | 21/284 [00:00<00:02, 102.78iter/s]
Build 3D deformation field: 11%|███▍ | 32/284 [00:00<00:02, 103.58iter/s]
Build 3D deformation field: 15%|████▋ | 43/284 [00:00<00:02, 104.01iter/s]
Build 3D deformation field: 19%|█████▉ | 54/284 [00:00<00:02, 105.72iter/s]
Build 3D deformation field: 23%|███████ | 65/284 [00:00<00:02, 105.76iter/s]
Build 3D deformation field: 27%|████████▎ | 76/284 [00:00<00:01, 105.57iter/s]
Build 3D deformation field: 31%|█████████▍ | 87/284 [00:00<00:01, 106.15iter/s]
Build 3D deformation field: 35%|██████████▋ | 98/284 [00:00<00:01, 106.01iter/s]
Build 3D deformation field: 38%|███████████▌ | 109/284 [00:01<00:01, 105.27iter/s]
Build 3D deformation field: 42%|████████████▋ | 120/284 [00:01<00:01, 105.14iter/s]
Build 3D deformation field: 46%|█████████████▊ | 131/284 [00:01<00:01, 104.96iter/s]
Build 3D deformation field: 50%|███████████████ | 142/284 [00:01<00:01, 105.17iter/s]
Build 3D deformation field: 54%|████████████████▏ | 153/284 [00:01<00:01, 105.11iter/s]
Build 3D deformation field: 58%|█████████████████▎ | 164/284 [00:01<00:01, 104.90iter/s]
Build 3D deformation field: 62%|██████████████████▍ | 175/284 [00:01<00:01, 105.07iter/s]
Build 3D deformation field: 65%|███████████████████▋ | 186/284 [00:01<00:00, 104.98iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 197/284 [00:01<00:00, 104.77iter/s]
Build 3D deformation field: 73%|█████████████████████▉ | 208/284 [00:01<00:00, 104.93iter/s]
Build 3D deformation field: 77%|███████████████████████▏ | 219/284 [00:02<00:00, 104.67iter/s]
Build 3D deformation field: 81%|████████████████████████▎ | 230/284 [00:02<00:00, 104.36iter/s]
Build 3D deformation field: 85%|█████████████████████████▍ | 241/284 [00:02<00:00, 104.35iter/s]
Build 3D deformation field: 89%|██████████████████████████▌ | 252/284 [00:02<00:00, 104.24iter/s]
Build 3D deformation field: 93%|███████████████████████████▊ | 263/284 [00:02<00:00, 105.40iter/s]
Build 3D deformation field: 96%|████████████████████████████▉ | 274/284 [00:02<00:00, 106.50iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 284/284 [00:02<00:00, 105.14iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
rm -rf /tmp/sct_2025-03-20_01-59-40_register-slicewise_0ktrmpdh
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx/warp_src2dest.nii.gz warp_sub-CRMBM3_acq-anat_TB1TFL2sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM3_acq-anat_TB1TFL2sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx/warp_dest2src.nii.gz warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_01-59-37_register-wrapper_zb_g6ilx
Finished! Elapsed time: 11s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015948.683280.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH1_acq-anat_TB1TFL.nii.gz -iseg sub-MGH1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MGH1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH1_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz (62, 118, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_01-59-53_register-slicewise_ck_86yo9)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 62 x 118 x 226
voxel size: 1.0mm x 1.0mm x 226mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/226 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/226 [00:00<00:48, 4.66iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
Estimate cord angle for each slice: 51%|███████████▎ | 116/226 [00:00<00:00, 461.89iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 226/226 [00:00<00:00, 554.00iter/s]
Build 3D deformation field: 0%| | 0/189 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▉ | 12/189 [00:00<00:01, 113.08iter/s]
Build 3D deformation field: 13%|███▉ | 24/189 [00:00<00:01, 115.82iter/s]
Build 3D deformation field: 19%|█████▉ | 36/189 [00:00<00:01, 116.38iter/s]
Build 3D deformation field: 26%|████████ | 49/189 [00:00<00:01, 118.55iter/s]
Build 3D deformation field: 33%|██████████▏ | 62/189 [00:00<00:01, 120.69iter/s]
Build 3D deformation field: 40%|████████████▎ | 75/189 [00:00<00:00, 120.65iter/s]
Build 3D deformation field: 47%|██████████████▍ | 88/189 [00:00<00:00, 121.02iter/s]
Build 3D deformation field: 53%|████████████████ | 101/189 [00:00<00:00, 121.90iter/s]
Build 3D deformation field: 60%|██████████████████ | 114/189 [00:00<00:00, 120.86iter/s]
Build 3D deformation field: 67%|████████████████████▏ | 127/189 [00:01<00:00, 120.31iter/s]
Build 3D deformation field: 74%|██████████████████████▏ | 140/189 [00:01<00:00, 120.16iter/s]
Build 3D deformation field: 81%|████████████████████████▎ | 153/189 [00:01<00:00, 120.96iter/s]
Build 3D deformation field: 88%|██████████████████████████▎ | 166/189 [00:01<00:00, 121.53iter/s]
Build 3D deformation field: 95%|████████████████████████████▍ | 179/189 [00:01<00:00, 122.06iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 189/189 [00:01<00:00, 120.60iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
rm -rf /tmp/sct_2025-03-20_01-59-53_register-slicewise_ck_86yo9
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24/warp_src2dest.nii.gz warp_sub-MGH1_acq-anat_TB1TFL2sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH1_acq-anat_TB1TFL2sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MGH1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24/warp_dest2src.nii.gz warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_01-59-52_register-wrapper_hmjvyn24
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_015958.850593.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH2_acq-anat_TB1TFL.nii.gz -iseg sub-MGH2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MGH2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH2_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz (59, 117, 223)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-03_register-slicewise_s2maqghw)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 59 x 117 x 223
voxel size: 1.0mm x 1.0mm x 223mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/223 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/223 [00:00<00:48, 4.60iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 54%|███████████▊ | 120/223 [00:00<00:00, 474.84iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 223/223 [00:00<00:00, 552.18iter/s]
Build 3D deformation field: 0%| | 0/218 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▋ | 12/218 [00:00<00:01, 116.50iter/s]
Build 3D deformation field: 11%|███▌ | 25/218 [00:00<00:01, 121.72iter/s]
Build 3D deformation field: 17%|█████▍ | 38/218 [00:00<00:01, 123.02iter/s]
Build 3D deformation field: 23%|███████▎ | 51/218 [00:00<00:01, 124.17iter/s]
Build 3D deformation field: 29%|█████████ | 64/218 [00:00<00:01, 124.69iter/s]
Build 3D deformation field: 35%|██████████▉ | 77/218 [00:00<00:01, 124.65iter/s]
Build 3D deformation field: 41%|████████████▊ | 90/218 [00:00<00:01, 125.43iter/s]
Build 3D deformation field: 47%|██████████████▏ | 103/218 [00:00<00:00, 125.81iter/s]
Build 3D deformation field: 53%|███████████████▉ | 116/218 [00:00<00:00, 123.52iter/s]
Build 3D deformation field: 59%|█████████████████▊ | 129/218 [00:01<00:00, 123.39iter/s]
Build 3D deformation field: 65%|███████████████████▌ | 142/218 [00:01<00:00, 124.54iter/s]
Build 3D deformation field: 71%|█████████████████████▎ | 155/218 [00:01<00:00, 126.12iter/s]
Build 3D deformation field: 77%|███████████████████████ | 168/218 [00:01<00:00, 125.30iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 181/218 [00:01<00:00, 126.15iter/s]
Build 3D deformation field: 89%|██████████████████████████▊ | 195/218 [00:01<00:00, 127.29iter/s]
Build 3D deformation field: 95%|████████████████████████████▌ | 208/218 [00:01<00:00, 127.55iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 218/218 [00:01<00:00, 125.45iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
rm -rf /tmp/sct_2025-03-20_02-00-03_register-slicewise_s2maqghw
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii/warp_src2dest.nii.gz warp_sub-MGH2_acq-anat_TB1TFL2sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH2_acq-anat_TB1TFL2sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MGH2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii/warp_dest2src.nii.gz warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-02_register-wrapper_21b5hkii
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020008.944421.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_acq-anat_TB1TFL.nii.gz -iseg sub-MGH3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MGH3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH3_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz (63, 127, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-14_register-slicewise_ue08b9db)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 63 x 127 x 238
voxel size: 1.0mm x 1.0mm x 238mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/238 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/238 [00:00<00:51, 4.60iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
Estimate cord angle for each slice: 49%|██████████▋ | 116/238 [00:00<00:00, 458.04iter/s]
Estimate cord angle for each slice: 99%|█████████████████████▋| 235/238 [00:00<00:00, 721.32iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #237 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 238/238 [00:00<00:00, 565.12iter/s]
Build 3D deformation field: 0%| | 0/216 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▌ | 11/216 [00:00<00:01, 106.46iter/s]
Build 3D deformation field: 11%|███▎ | 23/216 [00:00<00:01, 110.89iter/s]
Build 3D deformation field: 16%|█████ | 35/216 [00:00<00:01, 112.34iter/s]
Build 3D deformation field: 22%|██████▋ | 47/216 [00:00<00:01, 110.31iter/s]
Build 3D deformation field: 27%|████████▍ | 59/216 [00:00<00:01, 111.41iter/s]
Build 3D deformation field: 33%|██████████▏ | 71/216 [00:00<00:01, 112.20iter/s]
Build 3D deformation field: 38%|███████████▉ | 83/216 [00:00<00:01, 112.64iter/s]
Build 3D deformation field: 44%|█████████████▋ | 95/216 [00:00<00:01, 113.13iter/s]
Build 3D deformation field: 50%|██████████████▊ | 107/216 [00:00<00:00, 113.37iter/s]
Build 3D deformation field: 55%|████████████████▌ | 119/216 [00:01<00:00, 113.50iter/s]
Build 3D deformation field: 61%|██████████████████▏ | 131/216 [00:01<00:00, 113.56iter/s]
Build 3D deformation field: 66%|███████████████████▊ | 143/216 [00:01<00:00, 113.67iter/s]
Build 3D deformation field: 72%|█████████████████████▌ | 155/216 [00:01<00:00, 113.60iter/s]
Build 3D deformation field: 77%|███████████████████████▏ | 167/216 [00:01<00:00, 113.77iter/s]
Build 3D deformation field: 83%|████████████████████████▊ | 179/216 [00:01<00:00, 113.89iter/s]
Build 3D deformation field: 88%|██████████████████████████▌ | 191/216 [00:01<00:00, 114.01iter/s]
Build 3D deformation field: 94%|████████████████████████████▏ | 203/216 [00:01<00:00, 113.34iter/s]
Build 3D deformation field: 100%|█████████████████████████████▊| 215/216 [00:01<00:00, 113.45iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 216/216 [00:01<00:00, 112.96iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
rm -rf /tmp/sct_2025-03-20_02-00-14_register-slicewise_ue08b9db
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg/warp_src2dest.nii.gz warp_sub-MGH3_acq-anat_TB1TFL2sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH3_acq-anat_TB1TFL2sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MGH3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg/warp_dest2src.nii.gz warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-12_register-wrapper_v4nbkvfg
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020019.921665.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI1_acq-anat_TB1TFL.nii.gz -iseg sub-MNI1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MNI1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI1_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 77, 367)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-25_register-slicewise_bj53ncg7)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 77 x 367
voxel size: 0.7065217mm x 0.7000043mm x 367mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/367 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/367 [00:00<01:20, 4.55iter/s]
Estimate cord angle for each slice: 33%|███████▎ | 122/367 [00:00<00:00, 479.28iter/s]
Estimate cord angle for each slice: 67%|██████████████▋ | 245/367 [00:00<00:00, 749.53iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #364 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #365 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #366 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 367/367 [00:00<00:00, 707.32iter/s]
Build 3D deformation field: 0%| | 0/363 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▍ | 17/363 [00:00<00:02, 166.87iter/s]
Build 3D deformation field: 10%|██▉ | 35/363 [00:00<00:01, 174.00iter/s]
Build 3D deformation field: 15%|████▌ | 53/363 [00:00<00:01, 174.21iter/s]
Build 3D deformation field: 20%|██████ | 71/363 [00:00<00:01, 175.50iter/s]
Build 3D deformation field: 25%|███████▌ | 89/363 [00:00<00:01, 175.05iter/s]
Build 3D deformation field: 29%|████████▊ | 107/363 [00:00<00:01, 175.01iter/s]
Build 3D deformation field: 34%|██████████▎ | 125/363 [00:00<00:01, 175.33iter/s]
Build 3D deformation field: 39%|███████████▊ | 143/363 [00:00<00:01, 175.13iter/s]
Build 3D deformation field: 44%|█████████████▎ | 161/363 [00:00<00:01, 174.11iter/s]
Build 3D deformation field: 49%|██████████████▊ | 179/363 [00:01<00:01, 173.25iter/s]
Build 3D deformation field: 54%|████████████████▎ | 197/363 [00:01<00:00, 173.52iter/s]
Build 3D deformation field: 59%|█████████████████▊ | 215/363 [00:01<00:00, 173.35iter/s]
Build 3D deformation field: 64%|███████████████████▎ | 233/363 [00:01<00:00, 173.62iter/s]
Build 3D deformation field: 69%|████████████████████▋ | 251/363 [00:01<00:00, 173.85iter/s]
Build 3D deformation field: 74%|██████████████████████▏ | 269/363 [00:01<00:00, 174.67iter/s]
Build 3D deformation field: 79%|███████████████████████▊ | 288/363 [00:01<00:00, 176.51iter/s]
Build 3D deformation field: 84%|█████████████████████████▎ | 306/363 [00:01<00:00, 175.28iter/s]
Build 3D deformation field: 89%|██████████████████████████▊ | 324/363 [00:01<00:00, 175.69iter/s]
Build 3D deformation field: 94%|████████████████████████████▎ | 342/363 [00:01<00:00, 176.29iter/s]
Build 3D deformation field: 99%|█████████████████████████████▊| 360/363 [00:02<00:00, 175.88iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 363/363 [00:02<00:00, 174.81iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
rm -rf /tmp/sct_2025-03-20_02-00-25_register-slicewise_bj53ncg7
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3/warp_src2dest.nii.gz warp_sub-MNI1_acq-anat_TB1TFL2sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI1_acq-anat_TB1TFL2sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MNI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3/warp_dest2src.nii.gz warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-23_register-wrapper_rwcfnue3
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020031.897343.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI2_acq-anat_TB1TFL.nii.gz -iseg sub-MNI2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MNI2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI2_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz (63, 75, 361)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-36_register-slicewise_j3x_0zzp)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 63 x 75 x 361
voxel size: 0.7065217mm x 0.7000043mm x 361mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/361 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/361 [00:00<01:18, 4.61iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 34%|███████▍ | 122/361 [00:00<00:00, 482.28iter/s]
Estimate cord angle for each slice: 68%|███████████████ | 247/361 [00:00<00:00, 757.77iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #348 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #349 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #350 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #351 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #352 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #353 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #354 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #355 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #356 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #357 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #358 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #359 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #360 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 361/361 [00:00<00:00, 714.33iter/s]
Build 3D deformation field: 0%| | 0/345 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▌ | 18/345 [00:00<00:01, 177.22iter/s]
Build 3D deformation field: 11%|███▎ | 37/345 [00:00<00:01, 181.62iter/s]
Build 3D deformation field: 17%|█████ | 57/345 [00:00<00:01, 185.71iter/s]
Build 3D deformation field: 22%|██████▊ | 76/345 [00:00<00:01, 185.62iter/s]
Build 3D deformation field: 28%|████████▌ | 95/345 [00:00<00:01, 186.36iter/s]
Build 3D deformation field: 33%|█████████▉ | 114/345 [00:00<00:01, 185.12iter/s]
Build 3D deformation field: 39%|███████████▋ | 134/345 [00:00<00:01, 186.82iter/s]
Build 3D deformation field: 45%|█████████████▍ | 154/345 [00:00<00:01, 187.89iter/s]
Build 3D deformation field: 50%|███████████████ | 173/345 [00:00<00:00, 185.74iter/s]
Build 3D deformation field: 56%|████████████████▋ | 192/345 [00:01<00:00, 185.77iter/s]
Build 3D deformation field: 61%|██████████████████▎ | 211/345 [00:01<00:00, 185.50iter/s]
Build 3D deformation field: 67%|████████████████████ | 230/345 [00:01<00:00, 184.39iter/s]
Build 3D deformation field: 72%|█████████████████████▋ | 249/345 [00:01<00:00, 183.06iter/s]
Build 3D deformation field: 78%|███████████████████████▎ | 268/345 [00:01<00:00, 182.28iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 287/345 [00:01<00:00, 182.28iter/s]
Build 3D deformation field: 89%|██████████████████████████▌ | 306/345 [00:01<00:00, 183.46iter/s]
Build 3D deformation field: 94%|████████████████████████████▎ | 326/345 [00:01<00:00, 185.85iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 345/345 [00:01<00:00, 185.97iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 345/345 [00:01<00:00, 184.93iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
rm -rf /tmp/sct_2025-03-20_02-00-36_register-slicewise_j3x_0zzp
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96/warp_src2dest.nii.gz warp_sub-MNI2_acq-anat_TB1TFL2sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI2_acq-anat_TB1TFL2sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MNI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96/warp_dest2src.nii.gz warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-34_register-wrapper_ky7dnm96
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020042.844537.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_acq-anat_TB1TFL.nii.gz -iseg sub-MNI3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MNI3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI3_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 102, 260)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-47_register-slicewise_cz3w0yer)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 102 x 260
voxel size: 0.7065217mm x 0.6999958mm x 260mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/260 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/260 [00:00<00:55, 4.67iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
Estimate cord angle for each slice: 46%|██████████▏ | 120/260 [00:00<00:00, 478.11iter/s]
Estimate cord angle for each slice: 93%|████████████████████▌ | 243/260 [00:00<00:00, 750.96iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #257 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #258 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #259 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 260/260 [00:00<00:00, 605.33iter/s]
Build 3D deformation field: 0%| | 0/252 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▍ | 12/252 [00:00<00:02, 117.20iter/s]
Build 3D deformation field: 10%|███ | 25/252 [00:00<00:01, 122.10iter/s]
Build 3D deformation field: 15%|████▋ | 38/252 [00:00<00:01, 123.78iter/s]
Build 3D deformation field: 21%|██████▍ | 52/252 [00:00<00:01, 126.37iter/s]
Build 3D deformation field: 26%|███████▉ | 65/252 [00:00<00:01, 127.03iter/s]
Build 3D deformation field: 31%|█████████▋ | 79/252 [00:00<00:01, 128.04iter/s]
Build 3D deformation field: 37%|███████████▎ | 92/252 [00:00<00:01, 128.51iter/s]
Build 3D deformation field: 42%|████████████▌ | 105/252 [00:00<00:01, 128.74iter/s]
Build 3D deformation field: 47%|██████████████ | 118/252 [00:00<00:01, 128.96iter/s]
Build 3D deformation field: 52%|███████████████▌ | 131/252 [00:01<00:00, 129.07iter/s]
Build 3D deformation field: 57%|█████████████████▏ | 144/252 [00:01<00:00, 129.18iter/s]
Build 3D deformation field: 62%|██████████████████▋ | 157/252 [00:01<00:00, 129.14iter/s]
Build 3D deformation field: 67%|████████████████████▏ | 170/252 [00:01<00:00, 127.58iter/s]
Build 3D deformation field: 73%|█████████████████████▊ | 183/252 [00:01<00:00, 127.67iter/s]
Build 3D deformation field: 78%|███████████████████████▎ | 196/252 [00:01<00:00, 127.27iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 209/252 [00:01<00:00, 126.82iter/s]
Build 3D deformation field: 88%|██████████████████████████▌ | 223/252 [00:01<00:00, 127.87iter/s]
Build 3D deformation field: 94%|████████████████████████████ | 236/252 [00:01<00:00, 127.05iter/s]
Build 3D deformation field: 99%|█████████████████████████████▋| 249/252 [00:01<00:00, 126.49iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 252/252 [00:01<00:00, 127.29iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
rm -rf /tmp/sct_2025-03-20_02-00-47_register-slicewise_cz3w0yer
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj/warp_src2dest.nii.gz warp_sub-MNI3_acq-anat_TB1TFL2sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI3_acq-anat_TB1TFL2sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MNI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj/warp_dest2src.nii.gz warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-45_register-wrapper__brtdokj
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020053.630418.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI1_acq-anat_TB1TFL.nii.gz -iseg sub-MPI1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MPI1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI1_acq-anat_TB1TFL.nii.gz (90, 144, 56)
Destination ......... ../anat/sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 89, 266)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-00-58_register-slicewise_b2sscxq4)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 68 x 89 x 266
voxel size: 0.7065217mm x 0.7000045mm x 266mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/266 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/266 [00:00<00:58, 4.57iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #40 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #41 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #42 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #43 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #44 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #45 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #46 is empty. It will be ignored.
Estimate cord angle for each slice: 46%|██████████ | 122/266 [00:00<00:00, 479.79iter/s]
Estimate cord angle for each slice: 93%|████████████████████▍ | 247/266 [00:00<00:00, 755.41iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 266/266 [00:00<00:00, 611.31iter/s]
Build 3D deformation field: 0%| | 0/248 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▊ | 14/248 [00:00<00:01, 135.41iter/s]
Build 3D deformation field: 12%|███▋ | 29/248 [00:00<00:01, 139.95iter/s]
Build 3D deformation field: 18%|█████▌ | 44/248 [00:00<00:01, 142.49iter/s]
Build 3D deformation field: 24%|███████▍ | 59/248 [00:00<00:01, 144.99iter/s]
Build 3D deformation field: 30%|█████████▎ | 74/248 [00:00<00:01, 146.44iter/s]
Build 3D deformation field: 36%|███████████▏ | 89/248 [00:00<00:01, 147.03iter/s]
Build 3D deformation field: 42%|████████████▌ | 104/248 [00:00<00:00, 145.57iter/s]
Build 3D deformation field: 48%|██████████████▍ | 119/248 [00:00<00:00, 145.41iter/s]
Build 3D deformation field: 54%|████████████████▏ | 134/248 [00:00<00:00, 144.61iter/s]
Build 3D deformation field: 60%|██████████████████▏ | 150/248 [00:01<00:00, 146.37iter/s]
Build 3D deformation field: 67%|███████████████████▉ | 165/248 [00:01<00:00, 145.40iter/s]
Build 3D deformation field: 73%|█████████████████████▉ | 181/248 [00:01<00:00, 147.09iter/s]
Build 3D deformation field: 79%|███████████████████████▊ | 197/248 [00:01<00:00, 148.19iter/s]
Build 3D deformation field: 86%|█████████████████████████▊ | 213/248 [00:01<00:00, 148.87iter/s]
Build 3D deformation field: 92%|███████████████████████████▌ | 228/248 [00:01<00:00, 148.66iter/s]
Build 3D deformation field: 98%|█████████████████████████████▍| 243/248 [00:01<00:00, 148.71iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 248/248 [00:01<00:00, 146.44iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
rm -rf /tmp/sct_2025-03-20_02-00-58_register-slicewise_b2sscxq4
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy/warp_src2dest.nii.gz warp_sub-MPI1_acq-anat_TB1TFL2sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI1_acq-anat_TB1TFL2sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MPI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy/warp_dest2src.nii.gz warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-00-56_register-wrapper_wbkniapy
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020103.541276.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI2_acq-anat_TB1TFL.nii.gz -iseg sub-MPI2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MPI2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI2_acq-anat_TB1TFL.nii.gz (90, 144, 56)
Destination ......... ../anat/sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz (61, 89, 276)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-01-07_register-slicewise_tey91h__)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 61 x 89 x 276
voxel size: 0.7065217mm x 0.6999956mm x 276mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/276 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/276 [00:00<00:59, 4.59iter/s]
Estimate cord angle for each slice: 45%|█████████▉ | 124/276 [00:00<00:00, 489.43iter/s]
Estimate cord angle for each slice: 91%|███████████████████▉ | 250/276 [00:00<00:00, 765.45iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 276/276 [00:00<00:00, 626.91iter/s]
Build 3D deformation field: 0%| | 0/276 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▋ | 15/276 [00:00<00:01, 144.58iter/s]
Build 3D deformation field: 11%|███▍ | 31/276 [00:00<00:01, 148.22iter/s]
Build 3D deformation field: 17%|█████▎ | 47/276 [00:00<00:01, 149.74iter/s]
Build 3D deformation field: 23%|███████ | 63/276 [00:00<00:01, 150.02iter/s]
Build 3D deformation field: 29%|████████▊ | 79/276 [00:00<00:01, 152.60iter/s]
Build 3D deformation field: 34%|██████████▋ | 95/276 [00:00<00:01, 152.63iter/s]
Build 3D deformation field: 40%|████████████ | 111/276 [00:00<00:01, 152.74iter/s]
Build 3D deformation field: 46%|█████████████▊ | 127/276 [00:00<00:00, 153.25iter/s]
Build 3D deformation field: 52%|███████████████▌ | 143/276 [00:00<00:00, 152.60iter/s]
Build 3D deformation field: 58%|█████████████████▎ | 159/276 [00:01<00:00, 152.31iter/s]
Build 3D deformation field: 63%|███████████████████ | 175/276 [00:01<00:00, 152.62iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 191/276 [00:01<00:00, 152.24iter/s]
Build 3D deformation field: 75%|██████████████████████▌ | 207/276 [00:01<00:00, 151.97iter/s]
Build 3D deformation field: 81%|████████████████████████▏ | 223/276 [00:01<00:00, 151.38iter/s]
Build 3D deformation field: 87%|█████████████████████████▉ | 239/276 [00:01<00:00, 151.20iter/s]
Build 3D deformation field: 92%|███████████████████████████▋ | 255/276 [00:01<00:00, 150.85iter/s]
Build 3D deformation field: 98%|█████████████████████████████▍| 271/276 [00:01<00:00, 150.86iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 276/276 [00:01<00:00, 151.44iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
rm -rf /tmp/sct_2025-03-20_02-01-07_register-slicewise_tey91h__
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah/warp_src2dest.nii.gz warp_sub-MPI2_acq-anat_TB1TFL2sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI2_acq-anat_TB1TFL2sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MPI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah/warp_dest2src.nii.gz warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-06_register-wrapper_qu1lmzah
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020113.331718.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_acq-anat_TB1TFL.nii.gz -iseg sub-MPI3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MPI3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI3_acq-anat_TB1TFL.nii.gz (90, 144, 56)
Destination ......... ../anat/sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 117, 282)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-01-18_register-slicewise_qyq_j_7s)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 68 x 117 x 282
voxel size: 0.7065217mm x 0.7mm x 282mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/282 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/282 [00:00<01:01, 4.56iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
Estimate cord angle for each slice: 41%|████████▉ | 115/282 [00:00<00:00, 451.91iter/s]
Estimate cord angle for each slice: 83%|██████████████████▎ | 234/282 [00:00<00:00, 715.64iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 282/282 [00:00<00:00, 612.79iter/s]
Build 3D deformation field: 0%| | 0/263 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 11/263 [00:00<00:02, 102.86iter/s]
Build 3D deformation field: 9%|██▋ | 23/263 [00:00<00:02, 108.58iter/s]
Build 3D deformation field: 13%|████▏ | 35/263 [00:00<00:02, 109.94iter/s]
Build 3D deformation field: 18%|█████▌ | 47/263 [00:00<00:01, 110.72iter/s]
Build 3D deformation field: 22%|██████▉ | 59/263 [00:00<00:01, 111.92iter/s]
Build 3D deformation field: 27%|████████▎ | 71/263 [00:00<00:01, 112.51iter/s]
Build 3D deformation field: 32%|█████████▊ | 83/263 [00:00<00:01, 112.75iter/s]
Build 3D deformation field: 36%|███████████▏ | 95/263 [00:00<00:01, 112.79iter/s]
Build 3D deformation field: 41%|████████████▏ | 107/263 [00:00<00:01, 113.17iter/s]
Build 3D deformation field: 45%|█████████████▌ | 119/263 [00:01<00:01, 113.53iter/s]
Build 3D deformation field: 50%|██████████████▉ | 131/263 [00:01<00:01, 113.96iter/s]
Build 3D deformation field: 54%|████████████████▎ | 143/263 [00:01<00:01, 113.81iter/s]
Build 3D deformation field: 59%|█████████████████▋ | 155/263 [00:01<00:00, 113.56iter/s]
Build 3D deformation field: 63%|███████████████████ | 167/263 [00:01<00:00, 112.33iter/s]
Build 3D deformation field: 68%|████████████████████▍ | 179/263 [00:01<00:00, 112.87iter/s]
Build 3D deformation field: 73%|█████████████████████▊ | 191/263 [00:01<00:00, 113.38iter/s]
Build 3D deformation field: 77%|███████████████████████▏ | 203/263 [00:01<00:00, 113.15iter/s]
Build 3D deformation field: 82%|████████████████████████▌ | 215/263 [00:01<00:00, 112.29iter/s]
Build 3D deformation field: 86%|█████████████████████████▉ | 227/263 [00:02<00:00, 112.42iter/s]
Build 3D deformation field: 91%|███████████████████████████▎ | 239/263 [00:02<00:00, 112.30iter/s]
Build 3D deformation field: 95%|████████████████████████████▋ | 251/263 [00:02<00:00, 111.01iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 110.64iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 112.07iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
rm -rf /tmp/sct_2025-03-20_02-01-18_register-slicewise_qyq_j_7s
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3/warp_src2dest.nii.gz warp_sub-MPI3_acq-anat_TB1TFL2sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI3_acq-anat_TB1TFL2sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MPI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3/warp_dest2src.nii.gz warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-16_register-wrapper_8qyb19u3
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020125.360814.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM1_acq-famp_TB1TFL.nii.gz -d ../anat/sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -identity 1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM1_acq-famp_TB1TFL.nii.gz (88, 128, 56)
Destination ......... ../anat/sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 72, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM1_acq-famp_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30/warp_src2dest.nii.gz warp_sub-MSSM1_acq-famp_TB1TFL2sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM1_acq-famp_TB1TFL2sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MSSM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30/warp_dest2src.nii.gz warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz
File created: warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-28_register-wrapper_sk2zdy30
Finished! Elapsed time: 2s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020130.661424.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM2_acq-famp_TB1TFL.nii.gz -d ../anat/sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -identity 1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM2_acq-famp_TB1TFL.nii.gz (88, 128, 56)
Destination ......... ../anat/sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz (62, 74, 228)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM2_acq-famp_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3/warp_src2dest.nii.gz warp_sub-MSSM2_acq-famp_TB1TFL2sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM2_acq-famp_TB1TFL2sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MSSM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3/warp_dest2src.nii.gz warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz
File created: warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-33_register-wrapper_0436lqo3
Finished! Elapsed time: 2s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020135.134387.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_acq-famp_TB1TFL.nii.gz -d ../anat/sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -identity 1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM3_acq-famp_TB1TFL.nii.gz (88, 128, 56)
Destination ......... ../anat/sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 104, 289)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz # in /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM3_acq-famp_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_/warp_src2dest.nii.gz warp_sub-MSSM3_acq-famp_TB1TFL2sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM3_acq-famp_TB1TFL2sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-MSSM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_/warp_dest2src.nii.gz warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz
File created: warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-37_register-wrapper_v_l3cy1_
Finished! Elapsed time: 4s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020140.945325.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU1_acq-anat_TB1TFL.nii.gz -iseg sub-NTNU1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-NTNU1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU1_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 65, 296)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-01-45_register-slicewise_ri1tj46e)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 65 x 296
voxel size: 0.7065217mm x 0.6999944mm x 296mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/296 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/296 [00:00<01:03, 4.64iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #37 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #38 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #39 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #40 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #41 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #42 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #43 is empty. It will be ignored.
Estimate cord angle for each slice: 40%|████████▋ | 117/296 [00:00<00:00, 464.54iter/s]
Estimate cord angle for each slice: 81%|█████████████████▉ | 241/296 [00:00<00:00, 744.39iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #289 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #290 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #291 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #292 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #293 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #294 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #295 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 296/296 [00:00<00:00, 649.92iter/s]
Build 3D deformation field: 0%| | 0/240 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▍ | 19/240 [00:00<00:01, 186.38iter/s]
Build 3D deformation field: 17%|█████▎ | 41/240 [00:00<00:00, 200.59iter/s]
Build 3D deformation field: 26%|████████ | 62/240 [00:00<00:00, 204.11iter/s]
Build 3D deformation field: 35%|██████████▊ | 84/240 [00:00<00:00, 207.60iter/s]
Build 3D deformation field: 44%|█████████████▎ | 106/240 [00:00<00:00, 209.82iter/s]
Build 3D deformation field: 53%|████████████████ | 128/240 [00:00<00:00, 210.27iter/s]
Build 3D deformation field: 62%|██████████████████▊ | 150/240 [00:00<00:00, 209.65iter/s]
Build 3D deformation field: 71%|█████████████████████▍ | 171/240 [00:00<00:00, 207.43iter/s]
Build 3D deformation field: 80%|████████████████████████ | 192/240 [00:00<00:00, 206.16iter/s]
Build 3D deformation field: 89%|██████████████████████████▋ | 213/240 [00:01<00:00, 203.37iter/s]
Build 3D deformation field: 98%|█████████████████████████████▎| 234/240 [00:01<00:00, 203.35iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 240/240 [00:01<00:00, 205.24iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
rm -rf /tmp/sct_2025-03-20_02-01-45_register-slicewise_ri1tj46e
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3/warp_src2dest.nii.gz warp_sub-NTNU1_acq-anat_TB1TFL2sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU1_acq-anat_TB1TFL2sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-NTNU1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3/warp_dest2src.nii.gz warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-43_register-wrapper_301kodr3
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020149.857359.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU2_acq-anat_TB1TFL.nii.gz -iseg sub-NTNU2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-NTNU2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU2_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 71, 234)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-01-53_register-slicewise_g28v_9er)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 71 x 234
voxel size: 0.7065217mm x 0.6999992mm x 234mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/234 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/234 [00:00<00:50, 4.63iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
Estimate cord angle for each slice: 54%|███████████▊ | 126/234 [00:00<00:00, 498.92iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #230 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #231 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #232 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #233 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 234/234 [00:00<00:00, 575.41iter/s]
Build 3D deformation field: 0%| | 0/225 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▎ | 17/225 [00:00<00:01, 161.84iter/s]
Build 3D deformation field: 16%|████▊ | 35/225 [00:00<00:01, 167.48iter/s]
Build 3D deformation field: 24%|███████▎ | 53/225 [00:00<00:01, 169.82iter/s]
Build 3D deformation field: 32%|█████████▊ | 71/225 [00:00<00:00, 170.79iter/s]
Build 3D deformation field: 40%|████████████▎ | 89/225 [00:00<00:00, 172.13iter/s]
Build 3D deformation field: 48%|██████████████▎ | 107/225 [00:00<00:00, 172.97iter/s]
Build 3D deformation field: 56%|████████████████▋ | 125/225 [00:00<00:00, 173.77iter/s]
Build 3D deformation field: 64%|███████████████████▏ | 144/225 [00:00<00:00, 176.07iter/s]
Build 3D deformation field: 72%|█████████████████████▌ | 162/225 [00:00<00:00, 176.14iter/s]
Build 3D deformation field: 80%|████████████████████████ | 180/225 [00:01<00:00, 175.33iter/s]
Build 3D deformation field: 88%|██████████████████████████▍ | 198/225 [00:01<00:00, 176.07iter/s]
Build 3D deformation field: 96%|████████████████████████████▊ | 216/225 [00:01<00:00, 176.24iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 225/225 [00:01<00:00, 173.87iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
rm -rf /tmp/sct_2025-03-20_02-01-53_register-slicewise_g28v_9er
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt/warp_src2dest.nii.gz warp_sub-NTNU2_acq-anat_TB1TFL2sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU2_acq-anat_TB1TFL2sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-NTNU2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt/warp_dest2src.nii.gz warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-01-52_register-wrapper_iou199jt
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020157.984877.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_acq-anat_TB1TFL.nii.gz -iseg sub-NTNU3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-NTNU3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU3_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz (60, 103, 322)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-02-02_register-slicewise_f28t0sxh)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 60 x 103 x 322
voxel size: 0.7065217mm x 0.6999952mm x 322mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/322 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/322 [00:00<01:09, 4.63iter/s]
Estimate cord angle for each slice: 39%|████████▍ | 124/322 [00:00<00:00, 491.77iter/s]
Estimate cord angle for each slice: 78%|█████████████████ | 250/322 [00:00<00:00, 769.19iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 322/322 [00:00<00:00, 678.79iter/s]
Build 3D deformation field: 0%| | 0/322 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 14/322 [00:00<00:02, 136.14iter/s]
Build 3D deformation field: 9%|██▋ | 28/322 [00:00<00:02, 138.21iter/s]
Build 3D deformation field: 13%|████▏ | 43/322 [00:00<00:01, 140.20iter/s]
Build 3D deformation field: 18%|█████▌ | 58/322 [00:00<00:01, 140.05iter/s]
Build 3D deformation field: 23%|███████ | 73/322 [00:00<00:01, 139.63iter/s]
Build 3D deformation field: 27%|████████▍ | 87/322 [00:00<00:01, 139.48iter/s]
Build 3D deformation field: 31%|█████████▍ | 101/322 [00:00<00:01, 139.30iter/s]
Build 3D deformation field: 36%|██████████▊ | 116/322 [00:00<00:01, 140.46iter/s]
Build 3D deformation field: 41%|████████████▏ | 131/322 [00:00<00:01, 140.60iter/s]
Build 3D deformation field: 45%|█████████████▌ | 146/322 [00:01<00:01, 141.67iter/s]
Build 3D deformation field: 50%|███████████████ | 161/322 [00:01<00:01, 141.61iter/s]
Build 3D deformation field: 55%|████████████████▍ | 176/322 [00:01<00:01, 141.06iter/s]
Build 3D deformation field: 59%|█████████████████▊ | 191/322 [00:01<00:00, 141.08iter/s]
Build 3D deformation field: 64%|███████████████████▏ | 206/322 [00:01<00:00, 140.72iter/s]
Build 3D deformation field: 69%|████████████████████▌ | 221/322 [00:01<00:00, 140.64iter/s]
Build 3D deformation field: 73%|█████████████████████▉ | 236/322 [00:01<00:00, 140.59iter/s]
Build 3D deformation field: 78%|███████████████████████▍ | 251/322 [00:01<00:00, 140.02iter/s]
Build 3D deformation field: 83%|████████████████████████▊ | 266/322 [00:01<00:00, 140.71iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 281/322 [00:02<00:00, 140.83iter/s]
Build 3D deformation field: 92%|███████████████████████████▌ | 296/322 [00:02<00:00, 139.93iter/s]
Build 3D deformation field: 96%|████████████████████████████▉ | 310/322 [00:02<00:00, 139.77iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 322/322 [00:02<00:00, 140.19iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
rm -rf /tmp/sct_2025-03-20_02-02-02_register-slicewise_f28t0sxh
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1/warp_src2dest.nii.gz warp_sub-NTNU3_acq-anat_TB1TFL2sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU3_acq-anat_TB1TFL2sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-NTNU3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1/warp_dest2src.nii.gz warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-02-00_register-wrapper_awh25iz1
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020209.185611.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL1_acq-anat_TB1TFL.nii.gz -iseg sub-UCL1_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-UCL1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL1_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz (67, 105, 333)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-02-14_register-slicewise_mhfqdbj6)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 67 x 105 x 333
voxel size: 0.7065217mm x 0.7mm x 333mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/333 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/333 [00:00<01:12, 4.60iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
Estimate cord angle for each slice: 35%|███████▋ | 116/333 [00:00<00:00, 458.23iter/s]
Estimate cord angle for each slice: 71%|███████████████▌ | 236/333 [00:00<00:00, 725.80iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 333/333 [00:00<00:00, 669.23iter/s]
Build 3D deformation field: 0%| | 0/303 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▏ | 12/303 [00:00<00:02, 117.33iter/s]
Build 3D deformation field: 8%|██▌ | 25/303 [00:00<00:02, 122.15iter/s]
Build 3D deformation field: 13%|███▉ | 38/303 [00:00<00:02, 123.82iter/s]
Build 3D deformation field: 17%|█████▏ | 51/303 [00:00<00:02, 124.89iter/s]
Build 3D deformation field: 21%|██████▌ | 64/303 [00:00<00:01, 125.36iter/s]
Build 3D deformation field: 25%|███████▉ | 77/303 [00:00<00:01, 125.31iter/s]
Build 3D deformation field: 30%|█████████▏ | 90/303 [00:00<00:01, 126.01iter/s]
Build 3D deformation field: 34%|██████████▏ | 103/303 [00:00<00:01, 126.22iter/s]
Build 3D deformation field: 38%|███████████▍ | 116/303 [00:00<00:01, 126.63iter/s]
Build 3D deformation field: 43%|████████████▊ | 129/303 [00:01<00:01, 126.95iter/s]
Build 3D deformation field: 47%|██████████████ | 142/303 [00:01<00:01, 126.88iter/s]
Build 3D deformation field: 51%|███████████████▎ | 155/303 [00:01<00:01, 127.00iter/s]
Build 3D deformation field: 55%|████████████████▋ | 168/303 [00:01<00:01, 127.03iter/s]
Build 3D deformation field: 60%|█████████████████▉ | 181/303 [00:01<00:00, 126.09iter/s]
Build 3D deformation field: 64%|███████████████████▏ | 194/303 [00:01<00:00, 125.89iter/s]
Build 3D deformation field: 68%|████████████████████▍ | 207/303 [00:01<00:00, 125.93iter/s]
Build 3D deformation field: 73%|█████████████████████▊ | 220/303 [00:01<00:00, 126.22iter/s]
Build 3D deformation field: 77%|███████████████████████ | 233/303 [00:01<00:00, 126.46iter/s]
Build 3D deformation field: 81%|████████████████████████▎ | 246/303 [00:01<00:00, 126.67iter/s]
Build 3D deformation field: 85%|█████████████████████████▋ | 259/303 [00:02<00:00, 126.83iter/s]
Build 3D deformation field: 90%|██████████████████████████▉ | 272/303 [00:02<00:00, 127.18iter/s]
Build 3D deformation field: 94%|████████████████████████████▏ | 285/303 [00:02<00:00, 127.20iter/s]
Build 3D deformation field: 98%|█████████████████████████████▌| 298/303 [00:02<00:00, 127.09iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 303/303 [00:02<00:00, 126.20iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
rm -rf /tmp/sct_2025-03-20_02-02-14_register-slicewise_mhfqdbj6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL1_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo/warp_src2dest.nii.gz warp_sub-UCL1_acq-anat_TB1TFL2sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL1_acq-anat_TB1TFL2sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-UCL1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo/warp_dest2src.nii.gz warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-anat_TB1TFL.nii.gz
File created: warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-02-12_register-wrapper_egk5arpo
Finished! Elapsed time: 10s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020222.010181.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL2_acq-anat_TB1TFL.nii.gz -iseg sub-UCL2_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-UCL2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL2_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz (75, 110, 287)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-02-27_register-slicewise_27gleik7)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 75 x 110 x 287
voxel size: 0.7065217mm x 0.7000031mm x 287mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/287 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/287 [00:00<01:02, 4.56iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 41%|████████▉ | 117/287 [00:00<00:00, 459.15iter/s]
Estimate cord angle for each slice: 82%|█████████████████▉ | 234/287 [00:00<00:00, 713.38iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #265 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #266 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #267 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #268 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #269 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 287/287 [00:00<00:00, 628.74iter/s]
Build 3D deformation field: 0%| | 0/262 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▏ | 10/262 [00:00<00:02, 96.01iter/s]
Build 3D deformation field: 8%|██▍ | 21/262 [00:00<00:02, 102.50iter/s]
Build 3D deformation field: 12%|███▊ | 32/262 [00:00<00:02, 105.83iter/s]
Build 3D deformation field: 16%|█████ | 43/262 [00:00<00:02, 107.26iter/s]
Build 3D deformation field: 21%|██████▌ | 55/262 [00:00<00:01, 108.39iter/s]
Build 3D deformation field: 25%|███████▊ | 66/262 [00:00<00:01, 108.88iter/s]
Build 3D deformation field: 30%|█████████▏ | 78/262 [00:00<00:01, 109.47iter/s]
Build 3D deformation field: 34%|██████████▋ | 90/262 [00:00<00:01, 109.98iter/s]
Build 3D deformation field: 39%|███████████▋ | 102/262 [00:00<00:01, 110.32iter/s]
Build 3D deformation field: 44%|█████████████ | 114/262 [00:01<00:01, 110.06iter/s]
Build 3D deformation field: 48%|██████████████▍ | 126/262 [00:01<00:01, 110.03iter/s]
Build 3D deformation field: 53%|███████████████▊ | 138/262 [00:01<00:01, 109.76iter/s]
Build 3D deformation field: 57%|█████████████████ | 149/262 [00:01<00:01, 109.64iter/s]
Build 3D deformation field: 61%|██████████████████▎ | 160/262 [00:01<00:00, 109.10iter/s]
Build 3D deformation field: 65%|███████████████████▌ | 171/262 [00:01<00:00, 108.77iter/s]
Build 3D deformation field: 70%|████████████████████▉ | 183/262 [00:01<00:00, 109.33iter/s]
Build 3D deformation field: 74%|██████████████████████▎ | 195/262 [00:01<00:00, 109.59iter/s]
Build 3D deformation field: 79%|███████████████████████▌ | 206/262 [00:01<00:00, 109.16iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 218/262 [00:02<00:00, 109.54iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 229/262 [00:02<00:00, 109.67iter/s]
Build 3D deformation field: 92%|███████████████████████████▍ | 240/262 [00:02<00:00, 109.54iter/s]
Build 3D deformation field: 96%|████████████████████████████▋ | 251/262 [00:02<00:00, 109.45iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 262/262 [00:02<00:00, 109.48iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 262/262 [00:02<00:00, 108.98iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
rm -rf /tmp/sct_2025-03-20_02-02-27_register-slicewise_27gleik7
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL2_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2/warp_src2dest.nii.gz warp_sub-UCL2_acq-anat_TB1TFL2sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL2_acq-anat_TB1TFL2sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-UCL2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2/warp_dest2src.nii.gz warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-anat_TB1TFL.nii.gz
File created: warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-02-25_register-wrapper_s6l5nck2
Finished! Elapsed time: 10s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020234.875466.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_acq-anat_TB1TFL.nii.gz -iseg sub-UCL3_acq-anat_TB1TFL_seg.nii.gz -d ../anat/sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz -dseg ../anat/sub-UCL3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL3_acq-anat_TB1TFL.nii.gz (88, 144, 56)
Destination ......... ../anat/sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 115, 300)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-02-40_register-slicewise_ukp213o6)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 115 x 300
voxel size: 0.7065217mm x 0.7mm x 300mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/300 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/300 [00:00<01:04, 4.63iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
Estimate cord angle for each slice: 39%|████████▋ | 118/300 [00:00<00:00, 468.45iter/s]
Estimate cord angle for each slice: 79%|█████████████████▎ | 236/300 [00:00<00:00, 724.82iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #266 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #267 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #268 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #269 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #289 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #290 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #291 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #292 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #293 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #294 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #295 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #296 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #297 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #298 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #299 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 300/300 [00:00<00:00, 651.30iter/s]
Build 3D deformation field: 0%| | 0/260 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 11/260 [00:00<00:02, 107.08iter/s]
Build 3D deformation field: 9%|██▋ | 23/260 [00:00<00:02, 110.09iter/s]
Build 3D deformation field: 13%|████▏ | 35/260 [00:00<00:02, 111.99iter/s]
Build 3D deformation field: 18%|█████▌ | 47/260 [00:00<00:01, 112.32iter/s]
Build 3D deformation field: 23%|███████ | 59/260 [00:00<00:01, 111.90iter/s]
Build 3D deformation field: 27%|████████▍ | 71/260 [00:00<00:01, 111.49iter/s]
Build 3D deformation field: 32%|█████████▉ | 83/260 [00:00<00:01, 112.16iter/s]
Build 3D deformation field: 37%|███████████▎ | 95/260 [00:00<00:01, 112.13iter/s]
Build 3D deformation field: 41%|████████████▎ | 107/260 [00:00<00:01, 112.82iter/s]
Build 3D deformation field: 46%|█████████████▋ | 119/260 [00:01<00:01, 113.33iter/s]
Build 3D deformation field: 50%|███████████████ | 131/260 [00:01<00:01, 113.86iter/s]
Build 3D deformation field: 55%|████████████████▌ | 143/260 [00:01<00:01, 114.08iter/s]
Build 3D deformation field: 60%|█████████████████▉ | 155/260 [00:01<00:00, 114.36iter/s]
Build 3D deformation field: 64%|███████████████████▎ | 167/260 [00:01<00:00, 114.16iter/s]
Build 3D deformation field: 69%|████████████████████▋ | 179/260 [00:01<00:00, 114.34iter/s]
Build 3D deformation field: 73%|██████████████████████ | 191/260 [00:01<00:00, 114.09iter/s]
Build 3D deformation field: 78%|███████████████████████▍ | 203/260 [00:01<00:00, 114.18iter/s]
Build 3D deformation field: 83%|████████████████████████▊ | 215/260 [00:01<00:00, 113.43iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 227/260 [00:02<00:00, 112.65iter/s]
Build 3D deformation field: 92%|███████████████████████████▌ | 239/260 [00:02<00:00, 112.35iter/s]
Build 3D deformation field: 97%|████████████████████████████▉ | 251/260 [00:02<00:00, 112.02iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 260/260 [00:02<00:00, 112.72iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
rm -rf /tmp/sct_2025-03-20_02-02-40_register-slicewise_ukp213o6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL3_acq-anat_TB1TFL_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0/warp_src2dest.nii.gz warp_sub-UCL3_acq-anat_TB1TFL2sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL3_acq-anat_TB1TFL2sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: sub-UCL3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0/warp_dest2src.nii.gz warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-anat_TB1TFL.nii.gz
File created: warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-anat_TB1TFL.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-02-37_register-wrapper_k8w4u__0
Finished! Elapsed time: 10s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020247.611224.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
Register DREAM to MP2RAGE#
# Register DREAM data (B1+ mapping) to the MP2RAGE scan ⏳
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
!sct_register_multimodal -i {subject}_acq-famp_TB1DREAM.nii.gz -d ../anat/{subject}_UNIT1_crop.nii.gz -dseg ../anat/{subject}_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc "{path_qc}"
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-CRMBM1_UNIT1_crop.nii.gz -dseg ../anat/sub-CRMBM1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-CRMBM1_UNIT1_crop.nii.gz (65, 100, 314)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76/warp_src2dest.nii.gz warp_sub-CRMBM1_acq-famp_TB1DREAM2sub-CRMBM1_UNIT1_crop.nii.gz
File created: warp_sub-CRMBM1_acq-famp_TB1DREAM2sub-CRMBM1_UNIT1_crop.nii.gz
File created: sub-CRMBM1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76/warp_dest2src.nii.gz warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-02-50_register-wrapper_ed7lvw76
Finished! Elapsed time: 109s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020439.678778.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-CRMBM2_UNIT1_crop.nii.gz -dseg ../anat/sub-CRMBM2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-CRMBM2_UNIT1_crop.nii.gz (65, 90, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z/warp_src2dest.nii.gz warp_sub-CRMBM2_acq-famp_TB1DREAM2sub-CRMBM2_UNIT1_crop.nii.gz
File created: warp_sub-CRMBM2_acq-famp_TB1DREAM2sub-CRMBM2_UNIT1_crop.nii.gz
File created: sub-CRMBM2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z/warp_dest2src.nii.gz warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-04-42_register-wrapper_4004r6_z
Finished! Elapsed time: 85s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020607.149611.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-CRMBM3_UNIT1_crop.nii.gz -dseg ../anat/sub-CRMBM3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-CRMBM3_UNIT1_crop.nii.gz (69, 121, 337)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm/warp_src2dest.nii.gz warp_sub-CRMBM3_acq-famp_TB1DREAM2sub-CRMBM3_UNIT1_crop.nii.gz
File created: warp_sub-CRMBM3_acq-famp_TB1DREAM2sub-CRMBM3_UNIT1_crop.nii.gz
File created: sub-CRMBM3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm/warp_dest2src.nii.gz warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-06-09_register-wrapper_h0flp5wm
Finished! Elapsed time: 143s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020832.850609.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MGH1_UNIT1_crop.nii.gz -dseg ../anat/sub-MGH1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MGH1_UNIT1_crop.nii.gz (62, 118, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3/warp_src2dest.nii.gz warp_sub-MGH1_acq-famp_TB1DREAM2sub-MGH1_UNIT1_crop.nii.gz
File created: warp_sub-MGH1_acq-famp_TB1DREAM2sub-MGH1_UNIT1_crop.nii.gz
File created: sub-MGH1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3/warp_dest2src.nii.gz warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-08-36_register-wrapper_9bjzdfv3
Finished! Elapsed time: 70s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_020945.880704.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MGH2_UNIT1_crop.nii.gz -dseg ../anat/sub-MGH2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MGH2_UNIT1_crop.nii.gz (59, 117, 223)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw/warp_src2dest.nii.gz warp_sub-MGH2_acq-famp_TB1DREAM2sub-MGH2_UNIT1_crop.nii.gz
File created: warp_sub-MGH2_acq-famp_TB1DREAM2sub-MGH2_UNIT1_crop.nii.gz
File created: sub-MGH2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw/warp_dest2src.nii.gz warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-09-49_register-wrapper_kxnk8zsw
Finished! Elapsed time: 71s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021100.241644.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MGH3_UNIT1_crop.nii.gz -dseg ../anat/sub-MGH3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MGH3_UNIT1_crop.nii.gz (63, 127, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8/warp_src2dest.nii.gz warp_sub-MGH3_acq-famp_TB1DREAM2sub-MGH3_UNIT1_crop.nii.gz
File created: warp_sub-MGH3_acq-famp_TB1DREAM2sub-MGH3_UNIT1_crop.nii.gz
File created: sub-MGH3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8/warp_dest2src.nii.gz warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-11-03_register-wrapper_9hlosbc8
Finished! Elapsed time: 79s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021222.723117.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MNI1_UNIT1_crop.nii.gz -dseg ../anat/sub-MNI1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MNI1_UNIT1_crop.nii.gz (65, 77, 367)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0/warp_src2dest.nii.gz warp_sub-MNI1_acq-famp_TB1DREAM2sub-MNI1_UNIT1_crop.nii.gz
File created: warp_sub-MNI1_acq-famp_TB1DREAM2sub-MNI1_UNIT1_crop.nii.gz
File created: sub-MNI1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0/warp_dest2src.nii.gz warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-12-26_register-wrapper_8zl1r4o0
Finished! Elapsed time: 91s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021356.824557.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MNI2_UNIT1_crop.nii.gz -dseg ../anat/sub-MNI2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MNI2_UNIT1_crop.nii.gz (63, 75, 361)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd/warp_src2dest.nii.gz warp_sub-MNI2_acq-famp_TB1DREAM2sub-MNI2_UNIT1_crop.nii.gz
File created: warp_sub-MNI2_acq-famp_TB1DREAM2sub-MNI2_UNIT1_crop.nii.gz
File created: sub-MNI2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd/warp_dest2src.nii.gz warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-13-59_register-wrapper_khlsm1cd
Finished! Elapsed time: 89s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021528.403526.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MNI3_UNIT1_crop.nii.gz -dseg ../anat/sub-MNI3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MNI3_UNIT1_crop.nii.gz (69, 102, 260)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve/warp_src2dest.nii.gz warp_sub-MNI3_acq-famp_TB1DREAM2sub-MNI3_UNIT1_crop.nii.gz
File created: warp_sub-MNI3_acq-famp_TB1DREAM2sub-MNI3_UNIT1_crop.nii.gz
File created: sub-MNI3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve/warp_dest2src.nii.gz warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-15-31_register-wrapper_7sughrve
Finished! Elapsed time: 119s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021729.778231.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MPI1_UNIT1_crop.nii.gz -dseg ../anat/sub-MPI1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MPI1_UNIT1_crop.nii.gz (68, 89, 266)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj/warp_src2dest.nii.gz warp_sub-MPI1_acq-famp_TB1DREAM2sub-MPI1_UNIT1_crop.nii.gz
File created: warp_sub-MPI1_acq-famp_TB1DREAM2sub-MPI1_UNIT1_crop.nii.gz
File created: sub-MPI1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj/warp_dest2src.nii.gz warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-17-32_register-wrapper_g_ecovtj
Finished! Elapsed time: 98s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_021910.498083.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MPI2_UNIT1_crop.nii.gz -dseg ../anat/sub-MPI2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MPI2_UNIT1_crop.nii.gz (61, 89, 276)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_/warp_src2dest.nii.gz warp_sub-MPI2_acq-famp_TB1DREAM2sub-MPI2_UNIT1_crop.nii.gz
File created: warp_sub-MPI2_acq-famp_TB1DREAM2sub-MPI2_UNIT1_crop.nii.gz
File created: sub-MPI2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_/warp_dest2src.nii.gz warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-19-13_register-wrapper_u8_om8d_
Finished! Elapsed time: 88s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022041.129773.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MPI3_UNIT1_crop.nii.gz -dseg ../anat/sub-MPI3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MPI3_UNIT1_crop.nii.gz (68, 117, 282)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt/warp_src2dest.nii.gz warp_sub-MPI3_acq-famp_TB1DREAM2sub-MPI3_UNIT1_crop.nii.gz
File created: warp_sub-MPI3_acq-famp_TB1DREAM2sub-MPI3_UNIT1_crop.nii.gz
File created: sub-MPI3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt/warp_dest2src.nii.gz warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-20-43_register-wrapper_vwswswxt
Finished! Elapsed time: 143s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022307.105384.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MSSM1_UNIT1_crop.nii.gz -dseg ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MSSM1_UNIT1_crop.nii.gz (68, 72, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx/warp_src2dest.nii.gz warp_sub-MSSM1_acq-famp_TB1DREAM2sub-MSSM1_UNIT1_crop.nii.gz
File created: warp_sub-MSSM1_acq-famp_TB1DREAM2sub-MSSM1_UNIT1_crop.nii.gz
File created: sub-MSSM1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx/warp_dest2src.nii.gz warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-23-10_register-wrapper_m_0xc2dx
Finished! Elapsed time: 54s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022404.292391.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MSSM2_UNIT1_crop.nii.gz -dseg ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MSSM2_UNIT1_crop.nii.gz (62, 74, 228)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui/warp_src2dest.nii.gz warp_sub-MSSM2_acq-famp_TB1DREAM2sub-MSSM2_UNIT1_crop.nii.gz
File created: warp_sub-MSSM2_acq-famp_TB1DREAM2sub-MSSM2_UNIT1_crop.nii.gz
File created: sub-MSSM2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui/warp_dest2src.nii.gz warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-24-06_register-wrapper_17yhlqui
Finished! Elapsed time: 63s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022509.969471.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-MSSM3_UNIT1_crop.nii.gz -dseg ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-MSSM3_UNIT1_crop.nii.gz (65, 104, 289)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp/warp_src2dest.nii.gz warp_sub-MSSM3_acq-famp_TB1DREAM2sub-MSSM3_UNIT1_crop.nii.gz
File created: warp_sub-MSSM3_acq-famp_TB1DREAM2sub-MSSM3_UNIT1_crop.nii.gz
File created: sub-MSSM3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp/warp_dest2src.nii.gz warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-25-12_register-wrapper_t7vi5hxp
Finished! Elapsed time: 107s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022659.296890.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-NTNU1_UNIT1_crop.nii.gz -dseg ../anat/sub-NTNU1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-NTNU1_UNIT1_crop.nii.gz (65, 65, 296)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w/warp_src2dest.nii.gz warp_sub-NTNU1_acq-famp_TB1DREAM2sub-NTNU1_UNIT1_crop.nii.gz
File created: warp_sub-NTNU1_acq-famp_TB1DREAM2sub-NTNU1_UNIT1_crop.nii.gz
File created: sub-NTNU1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w/warp_dest2src.nii.gz warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-27-02_register-wrapper_e52svt9w
Finished! Elapsed time: 72s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022813.672538.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-NTNU2_UNIT1_crop.nii.gz -dseg ../anat/sub-NTNU2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-NTNU2_UNIT1_crop.nii.gz (69, 71, 234)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc/warp_src2dest.nii.gz warp_sub-NTNU2_acq-famp_TB1DREAM2sub-NTNU2_UNIT1_crop.nii.gz
File created: warp_sub-NTNU2_acq-famp_TB1DREAM2sub-NTNU2_UNIT1_crop.nii.gz
File created: sub-NTNU2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc/warp_dest2src.nii.gz warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-28-16_register-wrapper_0czdyagc
Finished! Elapsed time: 77s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_022933.531790.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-NTNU3_UNIT1_crop.nii.gz -dseg ../anat/sub-NTNU3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-NTNU3_UNIT1_crop.nii.gz (60, 103, 322)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2/warp_src2dest.nii.gz warp_sub-NTNU3_acq-famp_TB1DREAM2sub-NTNU3_UNIT1_crop.nii.gz
File created: warp_sub-NTNU3_acq-famp_TB1DREAM2sub-NTNU3_UNIT1_crop.nii.gz
File created: sub-NTNU3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2/warp_dest2src.nii.gz warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-29-35_register-wrapper_eswpgnb2
Finished! Elapsed time: 109s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023124.923140.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL1_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-UCL1_UNIT1_crop.nii.gz -dseg ../anat/sub-UCL1_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL1_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-UCL1_UNIT1_crop.nii.gz (67, 105, 333)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL1_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29/warp_src2dest.nii.gz warp_sub-UCL1_acq-famp_TB1DREAM2sub-UCL1_UNIT1_crop.nii.gz
File created: warp_sub-UCL1_acq-famp_TB1DREAM2sub-UCL1_UNIT1_crop.nii.gz
File created: sub-UCL1_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29/warp_dest2src.nii.gz warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-31-27_register-wrapper_y4x3sz29
Finished! Elapsed time: 120s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023328.239990.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL2_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-UCL2_UNIT1_crop.nii.gz -dseg ../anat/sub-UCL2_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL2_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-UCL2_UNIT1_crop.nii.gz (75, 110, 287)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL2_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt/warp_src2dest.nii.gz warp_sub-UCL2_acq-famp_TB1DREAM2sub-UCL2_UNIT1_crop.nii.gz
File created: warp_sub-UCL2_acq-famp_TB1DREAM2sub-UCL2_UNIT1_crop.nii.gz
File created: sub-UCL2_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt/warp_dest2src.nii.gz warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-33-31_register-wrapper_su8dndzt
Finished! Elapsed time: 112s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023523.610243.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_acq-famp_TB1DREAM.nii.gz -d ../anat/sub-UCL3_UNIT1_crop.nii.gz -dseg ../anat/sub-UCL3_UNIT1_seg_crop.nii.gz -param step=1,type=im,algo=slicereg,metric=CC,smooth=1 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL3_acq-famp_TB1DREAM.nii.gz (80, 80, 11)
Destination ......... ../anat/sub-UCL3_UNIT1_crop.nii.gz (69, 115, 300)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... im
algo ........... slicereg
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 1
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/runner/sct_6.5/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'CC[dest_RPI_crop.nii,src_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 1 -v 1 -o '[step1,src_reg_crop_regStep1.nii]' # in /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL3_acq-famp_TB1DREAM_reg.nii.gz
mv /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk/warp_src2dest.nii.gz warp_sub-UCL3_acq-famp_TB1DREAM2sub-UCL3_UNIT1_crop.nii.gz
File created: warp_sub-UCL3_acq-famp_TB1DREAM2sub-UCL3_UNIT1_crop.nii.gz
File created: sub-UCL3_UNIT1_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk/warp_dest2src.nii.gz warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz
File created: warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-35-26_register-wrapper_xykf59zk
Finished! Elapsed time: 135s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023741.523529.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
Segment spinal cord on SNR data#
# Segment spinal cord on SNR data
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
# Split SNR data into sub-volumes (https://github.com/spinal-cord-7t/coil-qc-code/issues/34)
!sct_image -i {subject}_acq-coilQaSagLarge_SNR.nii.gz -split t -o {subject}_acq-coilQaSagLarge_SNR.nii.gz
# Crop SNR data (https://github.com/spinal-cord-7t/coil-qc-code/issues/128)
!sct_crop_image -i {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz
!sct_crop_image -i {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -m {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -o {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz
# Segment spinal cord on SNR data, unless it already exists
fname_manual_seg = os.path.join(path_labels, subject, "fmap", f"{subject}_acq-coilQaSagLarge_SNR_T0000_label-SC_seg.nii.gz")
if os.path.exists(fname_manual_seg):
# Manual segmentation already exists. Copy it to local folder and put it in the same space as the SNR data
print(f"{subject}: Manual segmentation found\n")
!sct_register_multimodal -i {fname_manual_seg} -d {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -o {subject}_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -identity 1 -x linear
# Generate QC report to make sure the manual segmentation is correct
!sct_qc -i "{subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz" -s "{subject}_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz" -p sct_deepseg_sc -qc {path_qc} -qc-subject {subject}
else:
# Manual segmentation does not exist. Run automatic segmentation.
print(f"{subject}: Manual segmentation not found")
!sct_deepseg -i "{subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz" -task seg_sc_contrast_agnostic -thr 0 -qc {path_qc}
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-CRMBM1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-CRMBM1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-37-51_sct_deepseg_fzrmtf7m)
Copied sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-37-51_sct_deepseg_fzrmtf7m/sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 24 seconds
Saving results to: /tmp/sct_2025-03-20_02-37-51_sct_deepseg_fzrmtf7m/sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023815.859482.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-CRMBM2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-CRMBM2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-38-26_sct_deepseg_jtob1q9x)
Copied sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-38-26_sct_deepseg_jtob1q9x/sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-38-26_sct_deepseg_jtob1q9x/sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023850.983586.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-CRMBM3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-CRMBM3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-39-01_sct_deepseg_y59xv3zo)
Copied sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-39-01_sct_deepseg_y59xv3zo/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-39-01_sct_deepseg_y59xv3zo/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_023926.060570.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MGH1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MGH1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-39-37_sct_deepseg_n2l9lh7m)
Copied sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-39-37_sct_deepseg_n2l9lh7m/sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-39-37_sct_deepseg_n2l9lh7m/sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024001.373887.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MGH2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MGH2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-40-12_sct_deepseg_3dprrlfs)
Copied sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-40-12_sct_deepseg_3dprrlfs/sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-40-12_sct_deepseg_3dprrlfs/sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024036.550255.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MGH3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 383], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MGH3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-40-47_sct_deepseg_u5ed88q1)
Copied sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-40-47_sct_deepseg_u5ed88q1/sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-40-47_sct_deepseg_u5ed88q1/sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024111.781197.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MNI1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MNI1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-41-22_sct_deepseg_fnrqalle)
Copied sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-41-22_sct_deepseg_fnrqalle/sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-41-22_sct_deepseg_fnrqalle/sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024147.053144.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MNI2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MNI2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-41-57_sct_deepseg_k5_1qwnx)
Copied sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-41-57_sct_deepseg_k5_1qwnx/sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-41-57_sct_deepseg_k5_1qwnx/sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024222.072838.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MNI3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MNI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-42-33_sct_deepseg_4q7q9k43)
Copied sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-42-33_sct_deepseg_4q7q9k43/sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-42-33_sct_deepseg_4q7q9k43/sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024257.380186.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MPI1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MPI1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-43-08_sct_deepseg_b_wghe6c)
Copied sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-43-08_sct_deepseg_b_wghe6c/sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-43-08_sct_deepseg_b_wghe6c/sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024332.398693.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MPI2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[129, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MPI2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-43-43_sct_deepseg_ey29umxw)
Copied sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-43-43_sct_deepseg_ey29umxw/sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-43-43_sct_deepseg_ey29umxw/sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024407.428120.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MPI3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[129, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MPI3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-44-18_sct_deepseg_7id0w0wv)
Copied sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-44-18_sct_deepseg_7id0w0wv/sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-44-18_sct_deepseg_7id0w0wv/sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024442.519957.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MSSM1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[144, 361], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MSSM1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-44-53_sct_deepseg_qs_61fcq)
Copied sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-44-53_sct_deepseg_qs_61fcq/sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-44-53_sct_deepseg_qs_61fcq/sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024517.611422.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MSSM2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[172, 344], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MSSM2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-45-28_sct_deepseg_3gmi_x0u)
Copied sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-45-28_sct_deepseg_3gmi_x0u/sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 24 seconds
Saving results to: /tmp/sct_2025-03-20_02-45-28_sct_deepseg_3gmi_x0u/sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024552.435521.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-MSSM3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[154, 359], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-MSSM3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-46-02_sct_deepseg_14z_3ctr)
Copied sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-46-02_sct_deepseg_14z_3ctr/sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-46-02_sct_deepseg_14z_3ctr/sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024626.716313.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-NTNU1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-NTNU1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-46-37_sct_deepseg_xch5crhs)
Copied sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-46-37_sct_deepseg_xch5crhs/sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-46-37_sct_deepseg_xch5crhs/sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024701.481475.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-NTNU2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-NTNU2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-47-12_sct_deepseg_xu3kzr9i)
Copied sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-47-12_sct_deepseg_xu3kzr9i/sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-47-12_sct_deepseg_xu3kzr9i/sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024736.723436.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-NTNU3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-NTNU3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-47-47_sct_deepseg_d1fgf4or)
Copied sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-47-47_sct_deepseg_d1fgf4or/sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-47-47_sct_deepseg_d1fgf4or/sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024811.940152.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-UCL1_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-UCL1: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-48-22_sct_deepseg_ae0_szwz)
Copied sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-48-22_sct_deepseg_ae0_szwz/sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-48-22_sct_deepseg_ae0_szwz/sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024847.030510.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-UCL2_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-UCL2: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-48-57_sct_deepseg_sd7dh6my)
Copied sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-48-57_sct_deepseg_sd7dh6my/sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-48-57_sct_deepseg_sd7dh6my/sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024922.246672.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_image -i sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz -split t -o sub-UCL3_acq-coilQaSagLarge_SNR.nii.gz
--
Generate output files...
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -xmax 450 -o sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[0, 451], y=[0, 512], z=[0, 13]
Cropping the image...
File sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
--
Spinal Cord Toolbox (6.5)
sct_crop_image -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -m sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -o sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
--
Bounding box: x=[128, 384], y=[128, 384], z=[0, 13]
Cropping the image...
File sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz already exists. Will overwrite it.
sub-UCL3: Manual segmentation not found
--
Spinal Cord Toolbox (6.5)
sct_deepseg -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -task seg_sc_contrast_agnostic -thr 0 -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Model 'model_seg_sc_contrast_agnostic_softseg_monai' is up to date (Source: https://github.com/sct-pipeline/contrast-agnostic-softseg-spinalcord/releases/download/v2.5/model_contrast-agnostic_20240930-1002.zip)
Creating temporary folder (/tmp/sct_2025-03-20_02-49-33_sct_deepseg_kc87id_j)
Copied sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz to /tmp/sct_2025-03-20_02-49-33_sct_deepseg_kc87id_j/sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Starting inference...
Inference done.
Total inference time: 0 minute(s) 23 seconds
Saving results to: /tmp/sct_2025-03-20_02-49-33_sct_deepseg_kc87id_j/sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 vox
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_024957.311647.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
Register SNR to MP2RAGE#
# Register Rx coilQA data (SNR, g-factor) to the MP2RAGE scan ⏳
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
!sct_register_multimodal -i {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/{subject}_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg {subject}_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/{subject}_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc "{path_qc}"
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-CRMBM1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 100, 314)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-03_register-slicewise_5gf7pbtf)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 100 x 314
voxel size: 0.7065217mm x 0.6999998mm x 314mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/314 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/314 [00:00<01:07, 4.62iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1446: RuntimeWarning: invalid value encountered in divide
coordsrc /= coordsrc.std()
Slice #8 is empty. It will be ignored.
Estimate cord angle for each slice: 39%|████████▍ | 121/314 [00:00<00:00, 479.84iter/s]
Estimate cord angle for each slice: 78%|█████████████████▏ | 245/314 [00:00<00:00, 753.96iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #289 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #290 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #291 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #292 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #293 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #294 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #295 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #296 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #297 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #298 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #299 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #300 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #301 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #302 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #303 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #304 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #305 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #306 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #307 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #308 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #309 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #310 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #311 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #312 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #313 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 314/314 [00:00<00:00, 682.23iter/s]
Build 3D deformation field: 0%| | 0/261 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▌ | 13/261 [00:00<00:01, 124.94iter/s]
Build 3D deformation field: 10%|███▏ | 27/261 [00:00<00:01, 130.03iter/s]
Build 3D deformation field: 16%|████▊ | 41/261 [00:00<00:01, 131.37iter/s]
Build 3D deformation field: 21%|██████▌ | 55/261 [00:00<00:01, 132.26iter/s]
Build 3D deformation field: 26%|████████▏ | 69/261 [00:00<00:01, 132.26iter/s]
Build 3D deformation field: 32%|█████████▊ | 83/261 [00:00<00:01, 132.74iter/s]
Build 3D deformation field: 37%|███████████▌ | 97/261 [00:00<00:01, 132.47iter/s]
Build 3D deformation field: 43%|████████████▊ | 111/261 [00:00<00:01, 132.55iter/s]
Build 3D deformation field: 48%|██████████████▎ | 125/261 [00:00<00:01, 133.10iter/s]
Build 3D deformation field: 53%|███████████████▉ | 139/261 [00:01<00:00, 134.36iter/s]
Build 3D deformation field: 59%|█████████████████▌ | 153/261 [00:01<00:00, 133.92iter/s]
Build 3D deformation field: 64%|███████████████████▏ | 167/261 [00:01<00:00, 134.77iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 181/261 [00:01<00:00, 134.23iter/s]
Build 3D deformation field: 75%|██████████████████████▍ | 195/261 [00:01<00:00, 133.90iter/s]
Build 3D deformation field: 80%|████████████████████████ | 209/261 [00:01<00:00, 133.75iter/s]
Build 3D deformation field: 85%|█████████████████████████▋ | 223/261 [00:01<00:00, 132.86iter/s]
Build 3D deformation field: 91%|███████████████████████████▏ | 237/261 [00:01<00:00, 134.47iter/s]
Build 3D deformation field: 96%|████████████████████████████▊ | 251/261 [00:01<00:00, 134.03iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 261/261 [00:01<00:00, 133.34iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
rm -rf /tmp/sct_2025-03-20_02-50-03_register-slicewise_5gf7pbtf
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp/warp_src2dest.nii.gz warp_sub-CRMBM1_acq-coilQaSagLarge_SNR_T00002sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM1_acq-coilQaSagLarge_SNR_T00002sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp/warp_dest2src.nii.gz warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-01_register-wrapper_alqv6bjp
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025009.955670.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-CRMBM2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 90, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-14_register-slicewise_e6743vz4)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 90 x 226
voxel size: 0.7065217mm x 0.6999998mm x 226mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/226 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/226 [00:00<00:49, 4.53iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
Estimate cord angle for each slice: 54%|███████████▊ | 121/226 [00:00<00:00, 474.07iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #218 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #219 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #220 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #223 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #224 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #225 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 226/226 [00:00<00:00, 559.42iter/s]
Build 3D deformation field: 0%| | 0/214 [00:00<?, ?iter/s]
Build 3D deformation field: 7%|██▏ | 15/214 [00:00<00:01, 141.50iter/s]
Build 3D deformation field: 14%|████▎ | 30/214 [00:00<00:01, 143.75iter/s]
Build 3D deformation field: 21%|██████▌ | 45/214 [00:00<00:01, 144.97iter/s]
Build 3D deformation field: 28%|████████▋ | 60/214 [00:00<00:01, 145.64iter/s]
Build 3D deformation field: 35%|██████████▊ | 75/214 [00:00<00:00, 145.78iter/s]
Build 3D deformation field: 43%|█████████████▏ | 91/214 [00:00<00:00, 147.34iter/s]
Build 3D deformation field: 50%|██████████████▊ | 106/214 [00:00<00:00, 148.16iter/s]
Build 3D deformation field: 57%|█████████████████ | 122/214 [00:00<00:00, 150.00iter/s]
Build 3D deformation field: 64%|███████████████████▎ | 138/214 [00:00<00:00, 151.27iter/s]
Build 3D deformation field: 72%|█████████████████████▌ | 154/214 [00:01<00:00, 151.52iter/s]
Build 3D deformation field: 79%|███████████████████████▊ | 170/214 [00:01<00:00, 151.23iter/s]
Build 3D deformation field: 87%|██████████████████████████ | 186/214 [00:01<00:00, 151.74iter/s]
Build 3D deformation field: 94%|████████████████████████████▎ | 202/214 [00:01<00:00, 151.78iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 214/214 [00:01<00:00, 149.55iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
rm -rf /tmp/sct_2025-03-20_02-50-14_register-slicewise_e6743vz4
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g/warp_src2dest.nii.gz warp_sub-CRMBM2_acq-coilQaSagLarge_SNR_T00002sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM2_acq-coilQaSagLarge_SNR_T00002sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g/warp_dest2src.nii.gz warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-12_register-wrapper_3569et0g
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025018.989412.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-CRMBM3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 121, 337)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-24_register-slicewise_abvbccc6)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 121 x 337
voxel size: 0.7065217mm x 0.6999998mm x 337mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/337 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/337 [00:00<01:12, 4.64iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #37 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #38 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #39 is empty. It will be ignored.
Estimate cord angle for each slice: 33%|███████▎ | 112/337 [00:00<00:00, 444.79iter/s]
Estimate cord angle for each slice: 68%|██████████████▉ | 228/337 [00:00<00:00, 703.55iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #303 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #304 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #305 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #306 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #307 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #308 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #309 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #310 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #311 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #312 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #313 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #314 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #315 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #316 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #317 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #318 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #319 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #320 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #321 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #322 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #323 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #324 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #325 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #326 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #327 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #328 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #329 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #330 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #331 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #332 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #333 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #334 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #335 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #336 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 337/337 [00:00<00:00, 677.79iter/s]
Build 3D deformation field: 0%| | 0/263 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▏ | 10/263 [00:00<00:02, 95.33iter/s]
Build 3D deformation field: 8%|██▍ | 21/263 [00:00<00:02, 101.86iter/s]
Build 3D deformation field: 12%|███▊ | 32/263 [00:00<00:02, 104.45iter/s]
Build 3D deformation field: 16%|█████ | 43/263 [00:00<00:02, 105.19iter/s]
Build 3D deformation field: 21%|██████▎ | 54/263 [00:00<00:01, 105.54iter/s]
Build 3D deformation field: 25%|███████▋ | 65/263 [00:00<00:01, 106.23iter/s]
Build 3D deformation field: 29%|████████▉ | 76/263 [00:00<00:01, 107.29iter/s]
Build 3D deformation field: 33%|██████████▎ | 87/263 [00:00<00:01, 106.99iter/s]
Build 3D deformation field: 37%|███████████▌ | 98/263 [00:00<00:01, 107.42iter/s]
Build 3D deformation field: 41%|████████████▍ | 109/263 [00:01<00:01, 107.86iter/s]
Build 3D deformation field: 46%|█████████████▋ | 120/263 [00:01<00:01, 107.82iter/s]
Build 3D deformation field: 50%|██████████████▉ | 131/263 [00:01<00:01, 107.21iter/s]
Build 3D deformation field: 54%|████████████████▏ | 142/263 [00:01<00:01, 107.15iter/s]
Build 3D deformation field: 58%|█████████████████▍ | 153/263 [00:01<00:01, 106.24iter/s]
Build 3D deformation field: 62%|██████████████████▋ | 164/263 [00:01<00:00, 105.89iter/s]
Build 3D deformation field: 67%|███████████████████▉ | 175/263 [00:01<00:00, 104.91iter/s]
Build 3D deformation field: 71%|█████████████████████▏ | 186/263 [00:01<00:00, 105.29iter/s]
Build 3D deformation field: 75%|██████████████████████▍ | 197/263 [00:01<00:00, 106.03iter/s]
Build 3D deformation field: 79%|███████████████████████▋ | 208/263 [00:01<00:00, 106.47iter/s]
Build 3D deformation field: 83%|████████████████████████▉ | 219/263 [00:02<00:00, 107.35iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 230/263 [00:02<00:00, 108.05iter/s]
Build 3D deformation field: 92%|███████████████████████████▍ | 241/263 [00:02<00:00, 107.76iter/s]
Build 3D deformation field: 96%|████████████████████████████▋ | 252/263 [00:02<00:00, 108.09iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 108.12iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 106.62iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
rm -rf /tmp/sct_2025-03-20_02-50-24_register-slicewise_abvbccc6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw/warp_src2dest.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw/warp_dest2src.nii.gz warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-21_register-wrapper__8w7bpfw
Finished! Elapsed time: 11s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025032.382424.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MGH1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz (62, 118, 226)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-37_register-slicewise_xcdjumr6)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 62 x 118 x 226
voxel size: 1.0mm x 1.0mm x 226mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/226 [00:00<?, ?iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/226 [00:00<00:49, 4.56iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
Estimate cord angle for each slice: 55%|████████████▏ | 125/226 [00:00<00:00, 491.58iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 226/226 [00:00<00:00, 558.48iter/s]
Build 3D deformation field: 0%| | 0/189 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▉ | 12/189 [00:00<00:01, 113.48iter/s]
Build 3D deformation field: 13%|███▉ | 24/189 [00:00<00:01, 116.17iter/s]
Build 3D deformation field: 19%|█████▉ | 36/189 [00:00<00:01, 117.14iter/s]
Build 3D deformation field: 26%|████████ | 49/189 [00:00<00:01, 119.09iter/s]
Build 3D deformation field: 33%|██████████▏ | 62/189 [00:00<00:01, 119.29iter/s]
Build 3D deformation field: 40%|████████████▎ | 75/189 [00:00<00:00, 121.16iter/s]
Build 3D deformation field: 47%|██████████████▍ | 88/189 [00:00<00:00, 120.57iter/s]
Build 3D deformation field: 53%|████████████████ | 101/189 [00:00<00:00, 119.86iter/s]
Build 3D deformation field: 60%|██████████████████ | 114/189 [00:00<00:00, 120.76iter/s]
Build 3D deformation field: 67%|████████████████████▏ | 127/189 [00:01<00:00, 121.67iter/s]
Build 3D deformation field: 74%|██████████████████████▏ | 140/189 [00:01<00:00, 122.60iter/s]
Build 3D deformation field: 81%|████████████████████████▎ | 153/189 [00:01<00:00, 123.41iter/s]
Build 3D deformation field: 88%|██████████████████████████▎ | 166/189 [00:01<00:00, 122.98iter/s]
Build 3D deformation field: 95%|████████████████████████████▍ | 179/189 [00:01<00:00, 122.64iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 189/189 [00:01<00:00, 121.13iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
rm -rf /tmp/sct_2025-03-20_02-50-37_register-slicewise_xcdjumr6
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq/warp_src2dest.nii.gz warp_sub-MGH1_acq-coilQaSagLarge_SNR_T00002sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH1_acq-coilQaSagLarge_SNR_T00002sub-MGH1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MGH1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MGH1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq/warp_dest2src.nii.gz warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-35_register-wrapper_m5jx0zkq
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025042.252300.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MGH2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz (59, 117, 223)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-47_register-slicewise_35q8wiik)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 59 x 117 x 223
voxel size: 1.0mm x 1.0mm x 223mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/223 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/223 [00:00<00:49, 4.46iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 54%|███████████▉ | 121/223 [00:00<00:00, 469.06iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #211 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #212 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #213 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #214 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #215 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #216 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #217 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #218 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #219 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #220 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 223/223 [00:00<00:00, 550.33iter/s]
Build 3D deformation field: 0%| | 0/208 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▊ | 12/208 [00:00<00:01, 117.14iter/s]
Build 3D deformation field: 12%|███▋ | 25/208 [00:00<00:01, 121.43iter/s]
Build 3D deformation field: 18%|█████▋ | 38/208 [00:00<00:01, 122.63iter/s]
Build 3D deformation field: 25%|███████▌ | 51/208 [00:00<00:01, 123.61iter/s]
Build 3D deformation field: 31%|█████████▌ | 64/208 [00:00<00:01, 124.68iter/s]
Build 3D deformation field: 37%|███████████▍ | 77/208 [00:00<00:01, 125.68iter/s]
Build 3D deformation field: 43%|█████████████▍ | 90/208 [00:00<00:00, 125.49iter/s]
Build 3D deformation field: 50%|██████████████▊ | 103/208 [00:00<00:00, 125.37iter/s]
Build 3D deformation field: 56%|████████████████▋ | 116/208 [00:00<00:00, 124.45iter/s]
Build 3D deformation field: 62%|██████████████████▌ | 129/208 [00:01<00:00, 124.53iter/s]
Build 3D deformation field: 68%|████████████████████▍ | 142/208 [00:01<00:00, 125.24iter/s]
Build 3D deformation field: 75%|██████████████████████▌ | 156/208 [00:01<00:00, 127.01iter/s]
Build 3D deformation field: 81%|████████████████████████▍ | 169/208 [00:01<00:00, 126.36iter/s]
Build 3D deformation field: 88%|██████████████████████████▍ | 183/208 [00:01<00:00, 127.85iter/s]
Build 3D deformation field: 94%|████████████████████████████▎ | 196/208 [00:01<00:00, 128.38iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 208/208 [00:01<00:00, 126.03iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
rm -rf /tmp/sct_2025-03-20_02-50-47_register-slicewise_35q8wiik
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv/warp_src2dest.nii.gz warp_sub-MGH2_acq-coilQaSagLarge_SNR_T00002sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH2_acq-coilQaSagLarge_SNR_T00002sub-MGH2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MGH2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MGH2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv/warp_dest2src.nii.gz warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-45_register-wrapper_gtz_thmv
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025051.959963.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MGH3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz (255, 256, 13)
Destination ......... ../anat/sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz (63, 127, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-50-56_register-slicewise_ajstrew_)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 63 x 127 x 238
voxel size: 1.0mm x 1.0mm x 238mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/238 [00:00<?, ?iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/238 [00:00<00:51, 4.63iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
Estimate cord angle for each slice: 48%|██████████▌ | 114/238 [00:00<00:00, 452.17iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #180 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #181 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #200 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #201 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #202 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #203 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #204 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #205 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #206 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #230 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #231 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #232 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #233 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #234 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #235 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #236 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #237 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 238/238 [00:00<00:00, 578.20iter/s]
Build 3D deformation field: 0%| | 0/200 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▋ | 11/200 [00:00<00:01, 102.16iter/s]
Build 3D deformation field: 11%|███▍ | 22/200 [00:00<00:01, 105.37iter/s]
Build 3D deformation field: 16%|█████ | 33/200 [00:00<00:01, 106.78iter/s]
Build 3D deformation field: 22%|██████▉ | 45/200 [00:00<00:01, 108.30iter/s]
Build 3D deformation field: 28%|████████▊ | 57/200 [00:00<00:01, 108.93iter/s]
Build 3D deformation field: 34%|██████████▋ | 69/200 [00:00<00:01, 109.76iter/s]
Build 3D deformation field: 40%|████████████▍ | 80/200 [00:00<00:01, 109.55iter/s]
Build 3D deformation field: 46%|██████████████ | 91/200 [00:00<00:01, 108.80iter/s]
Build 3D deformation field: 51%|███████████████▎ | 102/200 [00:00<00:00, 108.22iter/s]
Build 3D deformation field: 56%|████████████████▉ | 113/200 [00:01<00:00, 107.66iter/s]
Build 3D deformation field: 62%|██████████████████▌ | 124/200 [00:01<00:00, 107.20iter/s]
Build 3D deformation field: 68%|████████████████████▎ | 135/200 [00:01<00:00, 107.70iter/s]
Build 3D deformation field: 73%|█████████████████████▉ | 146/200 [00:01<00:00, 106.98iter/s]
Build 3D deformation field: 78%|███████████████████████▌ | 157/200 [00:01<00:00, 106.99iter/s]
Build 3D deformation field: 84%|█████████████████████████▏ | 168/200 [00:01<00:00, 107.22iter/s]
Build 3D deformation field: 90%|██████████████████████████▊ | 179/200 [00:01<00:00, 107.83iter/s]
Build 3D deformation field: 96%|████████████████████████████▋ | 191/200 [00:01<00:00, 108.84iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 200/200 [00:01<00:00, 108.16iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
rm -rf /tmp/sct_2025-03-20_02-50-56_register-slicewise_ajstrew_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls/warp_src2dest.nii.gz warp_sub-MGH3_acq-coilQaSagLarge_SNR_T00002sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MGH3_acq-coilQaSagLarge_SNR_T00002sub-MGH3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MGH3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MGH3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls/warp_dest2src.nii.gz warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-50-55_register-wrapper_1tnw6dls
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025102.540817.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MNI1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 77, 367)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-08_register-slicewise_zmc4vnqo)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 77 x 367
voxel size: 0.7065217mm x 0.7000043mm x 367mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/367 [00:00<?, ?iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/367 [00:00<01:20, 4.57iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
Estimate cord angle for each slice: 35%|███████▋ | 128/367 [00:00<00:00, 503.32iter/s]
Estimate cord angle for each slice: 69%|███████████████ | 252/367 [00:00<00:00, 767.26iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #341 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #342 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #343 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #344 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #345 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #346 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #347 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #348 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #349 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #350 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #351 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #352 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #353 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #354 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #355 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #356 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #357 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #358 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #359 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #360 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #361 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #362 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #363 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #364 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #365 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #366 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 367/367 [00:00<00:00, 726.98iter/s]
Build 3D deformation field: 0%| | 0/320 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▋ | 17/320 [00:00<00:01, 162.07iter/s]
Build 3D deformation field: 11%|███▎ | 34/320 [00:00<00:01, 165.76iter/s]
Build 3D deformation field: 16%|█████ | 52/320 [00:00<00:01, 168.03iter/s]
Build 3D deformation field: 22%|██████▊ | 70/320 [00:00<00:01, 171.16iter/s]
Build 3D deformation field: 28%|████████▌ | 88/320 [00:00<00:01, 172.57iter/s]
Build 3D deformation field: 33%|█████████▉ | 106/320 [00:00<00:01, 173.10iter/s]
Build 3D deformation field: 39%|███████████▋ | 124/320 [00:00<00:01, 174.43iter/s]
Build 3D deformation field: 44%|█████████████▎ | 142/320 [00:00<00:01, 175.23iter/s]
Build 3D deformation field: 50%|███████████████ | 160/320 [00:00<00:00, 174.25iter/s]
Build 3D deformation field: 56%|████████████████▋ | 178/320 [00:01<00:00, 174.21iter/s]
Build 3D deformation field: 61%|██████████████████▍ | 196/320 [00:01<00:00, 172.24iter/s]
Build 3D deformation field: 67%|████████████████████ | 214/320 [00:01<00:00, 171.01iter/s]
Build 3D deformation field: 72%|█████████████████████▊ | 232/320 [00:01<00:00, 170.50iter/s]
Build 3D deformation field: 78%|███████████████████████▍ | 250/320 [00:01<00:00, 171.62iter/s]
Build 3D deformation field: 84%|█████████████████████████▏ | 268/320 [00:01<00:00, 171.74iter/s]
Build 3D deformation field: 89%|██████████████████████████▊ | 286/320 [00:01<00:00, 171.59iter/s]
Build 3D deformation field: 95%|████████████████████████████▌ | 304/320 [00:01<00:00, 172.16iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 320/320 [00:01<00:00, 172.12iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
rm -rf /tmp/sct_2025-03-20_02-51-08_register-slicewise_zmc4vnqo
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag/warp_src2dest.nii.gz warp_sub-MNI1_acq-coilQaSagLarge_SNR_T00002sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI1_acq-coilQaSagLarge_SNR_T00002sub-MNI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MNI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MNI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag/warp_dest2src.nii.gz warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-06_register-wrapper_592f4gag
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025114.235367.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MNI2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz (63, 75, 361)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-18_register-slicewise_8d7iitnt)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 63 x 75 x 361
voxel size: 0.7065217mm x 0.7000043mm x 361mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/361 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/361 [00:00<01:17, 4.63iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 34%|███████▌ | 124/361 [00:00<00:00, 491.61iter/s]
Estimate cord angle for each slice: 70%|███████████████▎ | 251/361 [00:00<00:00, 771.80iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #330 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #331 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #332 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #333 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #334 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #335 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #336 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #337 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #338 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #339 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #340 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #341 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #342 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #343 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #344 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #345 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #346 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #347 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #348 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #349 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #350 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #351 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #352 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #353 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #354 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #355 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #356 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #357 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #358 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #359 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #360 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 361/361 [00:00<00:00, 728.68iter/s]
Build 3D deformation field: 0%| | 0/327 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▋ | 18/327 [00:00<00:01, 178.58iter/s]
Build 3D deformation field: 11%|███▌ | 37/327 [00:00<00:01, 181.86iter/s]
Build 3D deformation field: 17%|█████▎ | 56/327 [00:00<00:01, 185.40iter/s]
Build 3D deformation field: 23%|███████▏ | 76/327 [00:00<00:01, 187.47iter/s]
Build 3D deformation field: 29%|█████████ | 95/327 [00:00<00:01, 188.08iter/s]
Build 3D deformation field: 35%|██████████▍ | 114/327 [00:00<00:01, 187.76iter/s]
Build 3D deformation field: 41%|████████████▏ | 133/327 [00:00<00:01, 187.77iter/s]
Build 3D deformation field: 46%|█████████████▉ | 152/327 [00:00<00:00, 188.27iter/s]
Build 3D deformation field: 53%|███████████████▊ | 172/327 [00:00<00:00, 189.11iter/s]
Build 3D deformation field: 58%|█████████████████▌ | 191/327 [00:01<00:00, 188.05iter/s]
Build 3D deformation field: 64%|███████████████████▎ | 210/327 [00:01<00:00, 186.58iter/s]
Build 3D deformation field: 70%|█████████████████████ | 229/327 [00:01<00:00, 187.22iter/s]
Build 3D deformation field: 76%|██████████████████████▊ | 248/327 [00:01<00:00, 187.72iter/s]
Build 3D deformation field: 82%|████████████████████████▍ | 267/327 [00:01<00:00, 188.19iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 286/327 [00:01<00:00, 187.62iter/s]
Build 3D deformation field: 93%|███████████████████████████▉ | 305/327 [00:01<00:00, 187.91iter/s]
Build 3D deformation field: 99%|█████████████████████████████▋| 324/327 [00:01<00:00, 187.83iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 327/327 [00:01<00:00, 187.39iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
rm -rf /tmp/sct_2025-03-20_02-51-18_register-slicewise_8d7iitnt
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z/warp_src2dest.nii.gz warp_sub-MNI2_acq-coilQaSagLarge_SNR_T00002sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI2_acq-coilQaSagLarge_SNR_T00002sub-MNI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MNI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MNI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z/warp_dest2src.nii.gz warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-17_register-wrapper__lveo86z
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025124.857550.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MNI3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 102, 260)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-29_register-slicewise_xxilkfvm)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 102 x 260
voxel size: 0.7065217mm x 0.6999958mm x 260mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/260 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/260 [00:00<00:56, 4.60iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
Estimate cord angle for each slice: 47%|██████████▏ | 121/260 [00:00<00:00, 478.57iter/s]
Estimate cord angle for each slice: 94%|████████████████████▋ | 244/260 [00:00<00:00, 748.87iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #257 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #258 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #259 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 260/260 [00:00<00:00, 602.02iter/s]
Build 3D deformation field: 0%| | 0/252 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▍ | 12/252 [00:00<00:02, 116.34iter/s]
Build 3D deformation field: 10%|███ | 25/252 [00:00<00:01, 120.50iter/s]
Build 3D deformation field: 15%|████▋ | 38/252 [00:00<00:01, 122.67iter/s]
Build 3D deformation field: 20%|██████▎ | 51/252 [00:00<00:01, 122.42iter/s]
Build 3D deformation field: 25%|███████▊ | 64/252 [00:00<00:01, 121.95iter/s]
Build 3D deformation field: 31%|█████████▍ | 77/252 [00:00<00:01, 122.99iter/s]
Build 3D deformation field: 36%|███████████ | 90/252 [00:00<00:01, 124.21iter/s]
Build 3D deformation field: 41%|████████████▎ | 103/252 [00:00<00:01, 125.17iter/s]
Build 3D deformation field: 46%|█████████████▊ | 116/252 [00:00<00:01, 125.70iter/s]
Build 3D deformation field: 51%|███████████████▎ | 129/252 [00:01<00:00, 126.05iter/s]
Build 3D deformation field: 56%|████████████████▉ | 142/252 [00:01<00:00, 125.68iter/s]
Build 3D deformation field: 62%|██████████████████▍ | 155/252 [00:01<00:00, 126.40iter/s]
Build 3D deformation field: 67%|████████████████████ | 168/252 [00:01<00:00, 126.43iter/s]
Build 3D deformation field: 72%|█████████████████████▌ | 181/252 [00:01<00:00, 127.22iter/s]
Build 3D deformation field: 77%|███████████████████████ | 194/252 [00:01<00:00, 127.36iter/s]
Build 3D deformation field: 82%|████████████████████████▋ | 207/252 [00:01<00:00, 127.02iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 220/252 [00:01<00:00, 127.61iter/s]
Build 3D deformation field: 92%|███████████████████████████▋ | 233/252 [00:01<00:00, 128.06iter/s]
Build 3D deformation field: 98%|█████████████████████████████▎| 246/252 [00:01<00:00, 127.72iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 252/252 [00:02<00:00, 125.73iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
rm -rf /tmp/sct_2025-03-20_02-51-29_register-slicewise_xxilkfvm
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg/warp_src2dest.nii.gz warp_sub-MNI3_acq-coilQaSagLarge_SNR_T00002sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MNI3_acq-coilQaSagLarge_SNR_T00002sub-MNI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MNI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MNI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg/warp_dest2src.nii.gz warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-27_register-wrapper_n9e67myg
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025135.547260.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MPI1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 89, 266)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-40_register-slicewise_1o3lvxqs)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 68 x 89 x 266
voxel size: 0.7065217mm x 0.7000045mm x 266mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/266 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/266 [00:00<00:58, 4.50iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #40 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #41 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #42 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #43 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #44 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #45 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #46 is empty. It will be ignored.
Estimate cord angle for each slice: 44%|█████████▊ | 118/266 [00:00<00:00, 459.52iter/s]
Estimate cord angle for each slice: 91%|███████████████████▉ | 241/266 [00:00<00:00, 733.61iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 266/266 [00:00<00:00, 598.93iter/s]
Build 3D deformation field: 0%| | 0/248 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▉ | 15/248 [00:00<00:01, 142.58iter/s]
Build 3D deformation field: 12%|███▊ | 30/248 [00:00<00:01, 145.41iter/s]
Build 3D deformation field: 18%|█████▋ | 45/248 [00:00<00:01, 143.44iter/s]
Build 3D deformation field: 24%|███████▌ | 60/248 [00:00<00:01, 144.33iter/s]
Build 3D deformation field: 30%|█████████▍ | 75/248 [00:00<00:01, 146.22iter/s]
Build 3D deformation field: 36%|███████████▎ | 90/248 [00:00<00:01, 147.37iter/s]
Build 3D deformation field: 42%|████████████▋ | 105/248 [00:00<00:00, 147.47iter/s]
Build 3D deformation field: 48%|██████████████▌ | 120/248 [00:00<00:00, 146.46iter/s]
Build 3D deformation field: 54%|████████████████▎ | 135/248 [00:00<00:00, 146.36iter/s]
Build 3D deformation field: 60%|██████████████████▏ | 150/248 [00:01<00:00, 145.04iter/s]
Build 3D deformation field: 67%|████████████████████ | 166/248 [00:01<00:00, 146.82iter/s]
Build 3D deformation field: 73%|█████████████████████▉ | 181/248 [00:01<00:00, 145.61iter/s]
Build 3D deformation field: 79%|███████████████████████▋ | 196/248 [00:01<00:00, 145.19iter/s]
Build 3D deformation field: 85%|█████████████████████████▋ | 212/248 [00:01<00:00, 146.71iter/s]
Build 3D deformation field: 92%|███████████████████████████▌ | 228/248 [00:01<00:00, 147.87iter/s]
Build 3D deformation field: 98%|█████████████████████████████▍| 243/248 [00:01<00:00, 148.17iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 248/248 [00:01<00:00, 146.55iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
rm -rf /tmp/sct_2025-03-20_02-51-40_register-slicewise_1o3lvxqs
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4/warp_src2dest.nii.gz warp_sub-MPI1_acq-coilQaSagLarge_SNR_T00002sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI1_acq-coilQaSagLarge_SNR_T00002sub-MPI1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MPI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MPI1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4/warp_dest2src.nii.gz warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-38_register-wrapper_f2bc1vt4
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025145.338428.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MPI2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz (255, 256, 13)
Destination ......... ../anat/sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz (61, 89, 276)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-49_register-slicewise_22oile17)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 61 x 89 x 276
voxel size: 0.7065217mm x 0.6999956mm x 276mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/276 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/276 [00:00<00:59, 4.61iter/s]
Estimate cord angle for each slice: 45%|█████████▊ | 123/276 [00:00<00:00, 486.16iter/s]
Estimate cord angle for each slice: 89%|███████████████████▋ | 247/276 [00:00<00:00, 757.00iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 276/276 [00:00<00:00, 623.66iter/s]
Build 3D deformation field: 0%| | 0/276 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▋ | 15/276 [00:00<00:01, 146.19iter/s]
Build 3D deformation field: 11%|███▍ | 31/276 [00:00<00:01, 152.70iter/s]
Build 3D deformation field: 17%|█████▎ | 47/276 [00:00<00:01, 153.95iter/s]
Build 3D deformation field: 23%|███████ | 63/276 [00:00<00:01, 155.95iter/s]
Build 3D deformation field: 29%|████████▊ | 79/276 [00:00<00:01, 156.12iter/s]
Build 3D deformation field: 34%|██████████▋ | 95/276 [00:00<00:01, 156.13iter/s]
Build 3D deformation field: 40%|████████████ | 111/276 [00:00<00:01, 156.85iter/s]
Build 3D deformation field: 46%|█████████████▊ | 127/276 [00:00<00:00, 157.33iter/s]
Build 3D deformation field: 52%|███████████████▌ | 143/276 [00:00<00:00, 156.98iter/s]
Build 3D deformation field: 58%|█████████████████▎ | 159/276 [00:01<00:00, 157.21iter/s]
Build 3D deformation field: 63%|███████████████████ | 175/276 [00:01<00:00, 157.61iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 191/276 [00:01<00:00, 157.65iter/s]
Build 3D deformation field: 75%|██████████████████████▌ | 207/276 [00:01<00:00, 157.39iter/s]
Build 3D deformation field: 81%|████████████████████████▏ | 223/276 [00:01<00:00, 156.98iter/s]
Build 3D deformation field: 87%|██████████████████████████ | 240/276 [00:01<00:00, 158.27iter/s]
Build 3D deformation field: 93%|███████████████████████████▉ | 257/276 [00:01<00:00, 159.93iter/s]
Build 3D deformation field: 99%|█████████████████████████████▋| 273/276 [00:01<00:00, 159.48iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 276/276 [00:01<00:00, 157.34iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
rm -rf /tmp/sct_2025-03-20_02-51-49_register-slicewise_22oile17
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe/warp_src2dest.nii.gz warp_sub-MPI2_acq-coilQaSagLarge_SNR_T00002sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI2_acq-coilQaSagLarge_SNR_T00002sub-MPI2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MPI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MPI2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe/warp_dest2src.nii.gz warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-48_register-wrapper_d1hpv8oe
Finished! Elapsed time: 7s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025154.887149.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MPI3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz (255, 256, 13)
Destination ......... ../anat/sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 117, 282)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-51-59_register-slicewise_oi52jkjv)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 68 x 117 x 282
voxel size: 0.7065217mm x 0.7mm x 282mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/282 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/282 [00:00<01:00, 4.66iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
Estimate cord angle for each slice: 42%|█████████▏ | 118/282 [00:00<00:00, 470.17iter/s]
Estimate cord angle for each slice: 85%|██████████████████▋ | 239/282 [00:00<00:00, 738.06iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 282/282 [00:00<00:00, 625.96iter/s]
Build 3D deformation field: 0%| | 0/263 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 11/263 [00:00<00:02, 106.18iter/s]
Build 3D deformation field: 9%|██▋ | 23/263 [00:00<00:02, 111.22iter/s]
Build 3D deformation field: 13%|████▏ | 35/263 [00:00<00:02, 112.65iter/s]
Build 3D deformation field: 18%|█████▌ | 47/263 [00:00<00:01, 113.48iter/s]
Build 3D deformation field: 22%|██████▉ | 59/263 [00:00<00:01, 113.89iter/s]
Build 3D deformation field: 27%|████████▎ | 71/263 [00:00<00:01, 114.00iter/s]
Build 3D deformation field: 32%|█████████▊ | 83/263 [00:00<00:01, 113.12iter/s]
Build 3D deformation field: 36%|███████████▏ | 95/263 [00:00<00:01, 113.14iter/s]
Build 3D deformation field: 41%|████████████▏ | 107/263 [00:00<00:01, 113.72iter/s]
Build 3D deformation field: 45%|█████████████▌ | 119/263 [00:01<00:01, 113.80iter/s]
Build 3D deformation field: 50%|██████████████▉ | 131/263 [00:01<00:01, 114.00iter/s]
Build 3D deformation field: 54%|████████████████▎ | 143/263 [00:01<00:01, 113.92iter/s]
Build 3D deformation field: 59%|█████████████████▋ | 155/263 [00:01<00:00, 114.07iter/s]
Build 3D deformation field: 63%|███████████████████ | 167/263 [00:01<00:00, 113.86iter/s]
Build 3D deformation field: 68%|████████████████████▍ | 179/263 [00:01<00:00, 113.30iter/s]
Build 3D deformation field: 73%|█████████████████████▊ | 191/263 [00:01<00:00, 112.73iter/s]
Build 3D deformation field: 77%|███████████████████████▏ | 203/263 [00:01<00:00, 112.69iter/s]
Build 3D deformation field: 82%|████████████████████████▌ | 215/263 [00:01<00:00, 113.16iter/s]
Build 3D deformation field: 86%|█████████████████████████▉ | 227/263 [00:02<00:00, 113.11iter/s]
Build 3D deformation field: 91%|███████████████████████████▎ | 239/263 [00:02<00:00, 112.28iter/s]
Build 3D deformation field: 95%|████████████████████████████▋ | 251/263 [00:02<00:00, 111.88iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 111.51iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 263/263 [00:02<00:00, 112.87iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
rm -rf /tmp/sct_2025-03-20_02-51-59_register-slicewise_oi52jkjv
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t/warp_src2dest.nii.gz warp_sub-MPI3_acq-coilQaSagLarge_SNR_T00002sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MPI3_acq-coilQaSagLarge_SNR_T00002sub-MPI3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MPI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MPI3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t/warp_dest2src.nii.gz warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-51-57_register-wrapper_ovybk15t
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025206.542359.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz (217, 256, 13)
Destination ......... ../anat/sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz (68, 72, 238)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-10_register-slicewise_xcjwu753)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 68 x 72 x 238
voxel size: 0.7065217mm x 0.7000028mm x 238mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/238 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/238 [00:00<00:51, 4.59iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
Estimate cord angle for each slice: 50%|███████████ | 119/238 [00:00<00:00, 469.64iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #233 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #234 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #235 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #236 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #237 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 238/238 [00:00<00:00, 570.26iter/s]
Build 3D deformation field: 0%| | 0/220 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▍ | 17/220 [00:00<00:01, 169.39iter/s]
Build 3D deformation field: 16%|████▉ | 35/220 [00:00<00:01, 173.57iter/s]
Build 3D deformation field: 24%|███████▍ | 53/220 [00:00<00:00, 175.68iter/s]
Build 3D deformation field: 33%|██████████▏ | 72/220 [00:00<00:00, 178.08iter/s]
Build 3D deformation field: 41%|████████████▊ | 91/220 [00:00<00:00, 179.51iter/s]
Build 3D deformation field: 50%|███████████████ | 110/220 [00:00<00:00, 180.89iter/s]
Build 3D deformation field: 59%|█████████████████▌ | 129/220 [00:00<00:00, 180.00iter/s]
Build 3D deformation field: 67%|████████████████████▏ | 148/220 [00:00<00:00, 179.99iter/s]
Build 3D deformation field: 76%|██████████████████████▊ | 167/220 [00:00<00:00, 180.54iter/s]
Build 3D deformation field: 85%|█████████████████████████▎ | 186/220 [00:01<00:00, 181.89iter/s]
Build 3D deformation field: 93%|███████████████████████████▉ | 205/220 [00:01<00:00, 182.73iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 220/220 [00:01<00:00, 180.35iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
rm -rf /tmp/sct_2025-03-20_02-52-10_register-slicewise_xcjwu753
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7/warp_src2dest.nii.gz warp_sub-MSSM1_acq-coilQaSagLarge_SNR_T00002sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM1_acq-coilQaSagLarge_SNR_T00002sub-MSSM1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MSSM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7/warp_dest2src.nii.gz warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-09_register-wrapper_9bnqtap7
Finished! Elapsed time: 5s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025214.945519.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz (172, 256, 13)
Destination ......... ../anat/sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz (62, 74, 228)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-18_register-slicewise_cc214bk9)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 62 x 74 x 228
voxel size: 0.7065217mm x 0.7000019mm x 228mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/228 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/228 [00:00<00:48, 4.65iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
Estimate cord angle for each slice: 54%|███████████▊ | 123/228 [00:00<00:00, 488.87iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #218 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #219 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #220 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #223 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #224 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #225 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #226 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #227 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 228/228 [00:00<00:00, 575.11iter/s]
Build 3D deformation field: 0%| | 0/213 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▌ | 18/213 [00:00<00:01, 174.75iter/s]
Build 3D deformation field: 17%|█████▍ | 37/213 [00:00<00:00, 181.95iter/s]
Build 3D deformation field: 26%|████████▏ | 56/213 [00:00<00:00, 185.47iter/s]
Build 3D deformation field: 35%|██████████▉ | 75/213 [00:00<00:00, 183.46iter/s]
Build 3D deformation field: 44%|█████████████▋ | 94/213 [00:00<00:00, 185.59iter/s]
Build 3D deformation field: 54%|████████████████ | 114/213 [00:00<00:00, 189.34iter/s]
Build 3D deformation field: 63%|██████████████████▊ | 134/213 [00:00<00:00, 190.01iter/s]
Build 3D deformation field: 72%|█████████████████████▋ | 154/213 [00:00<00:00, 191.75iter/s]
Build 3D deformation field: 82%|████████████████████████▌ | 174/213 [00:00<00:00, 192.61iter/s]
Build 3D deformation field: 91%|███████████████████████████▎ | 194/213 [00:01<00:00, 193.61iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 213/213 [00:01<00:00, 189.26iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
rm -rf /tmp/sct_2025-03-20_02-52-18_register-slicewise_cc214bk9
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w/warp_src2dest.nii.gz warp_sub-MSSM2_acq-coilQaSagLarge_SNR_T00002sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM2_acq-coilQaSagLarge_SNR_T00002sub-MSSM2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MSSM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w/warp_dest2src.nii.gz warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-17_register-wrapper_dtkcgb5w
Finished! Elapsed time: 5s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025222.285226.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (205, 256, 13)
Destination ......... ../anat/sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 104, 289)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-26_register-slicewise_0k5q6ecc)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 104 x 289
voxel size: 0.7065217mm x 0.7000019mm x 289mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/289 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/289 [00:00<01:01, 4.69iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
Estimate cord angle for each slice: 40%|████████▊ | 116/289 [00:00<00:00, 463.75iter/s]
Estimate cord angle for each slice: 81%|█████████████████▉ | 235/289 [00:00<00:00, 726.38iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #242 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #243 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #244 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #245 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #246 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #247 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #248 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #249 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #250 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #251 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #252 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #253 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #254 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #255 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #256 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #257 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #258 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #259 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #260 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #261 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #262 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #263 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #264 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #265 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #266 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #267 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #268 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #269 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 289/289 [00:00<00:00, 654.79iter/s]
Build 3D deformation field: 0%| | 0/218 [00:00<?, ?iter/s]
Build 3D deformation field: 6%|█▋ | 12/218 [00:00<00:01, 118.32iter/s]
Build 3D deformation field: 12%|███▋ | 26/218 [00:00<00:01, 126.31iter/s]
Build 3D deformation field: 18%|█████▌ | 39/218 [00:00<00:01, 127.89iter/s]
Build 3D deformation field: 24%|███████▌ | 53/218 [00:00<00:01, 130.12iter/s]
Build 3D deformation field: 31%|█████████▌ | 67/218 [00:00<00:01, 131.73iter/s]
Build 3D deformation field: 37%|███████████▌ | 81/218 [00:00<00:01, 131.76iter/s]
Build 3D deformation field: 44%|█████████████▌ | 95/218 [00:00<00:00, 132.21iter/s]
Build 3D deformation field: 50%|███████████████ | 109/218 [00:00<00:00, 132.84iter/s]
Build 3D deformation field: 56%|████████████████▉ | 123/218 [00:00<00:00, 131.46iter/s]
Build 3D deformation field: 63%|██████████████████▊ | 137/218 [00:01<00:00, 132.23iter/s]
Build 3D deformation field: 69%|████████████████████▊ | 151/218 [00:01<00:00, 131.63iter/s]
Build 3D deformation field: 76%|██████████████████████▋ | 165/218 [00:01<00:00, 131.85iter/s]
Build 3D deformation field: 82%|████████████████████████▋ | 179/218 [00:01<00:00, 131.83iter/s]
Build 3D deformation field: 89%|██████████████████████████▌ | 193/218 [00:01<00:00, 131.82iter/s]
Build 3D deformation field: 95%|████████████████████████████▍ | 207/218 [00:01<00:00, 132.65iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 218/218 [00:01<00:00, 131.35iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
rm -rf /tmp/sct_2025-03-20_02-52-26_register-slicewise_0k5q6ecc
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9/warp_src2dest.nii.gz warp_sub-MSSM3_acq-coilQaSagLarge_SNR_T00002sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-MSSM3_acq-coilQaSagLarge_SNR_T00002sub-MSSM3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-MSSM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9/warp_dest2src.nii.gz warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-24_register-wrapper_zh7wazy9
Finished! Elapsed time: 8s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025232.244516.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-NTNU1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz (65, 65, 296)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-36_register-slicewise_uyymd0dk)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 65 x 65 x 296
voxel size: 0.7065217mm x 0.6999944mm x 296mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/296 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/296 [00:00<01:03, 4.66iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #37 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #38 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #39 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #40 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #41 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #42 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #43 is empty. It will be ignored.
Estimate cord angle for each slice: 41%|████████▉ | 120/296 [00:00<00:00, 477.14iter/s]
Estimate cord angle for each slice: 82%|██████████████████▏ | 244/296 [00:00<00:00, 753.68iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #289 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #290 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #291 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #292 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #293 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #294 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #295 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 296/296 [00:00<00:00, 653.90iter/s]
Build 3D deformation field: 0%| | 0/238 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▌ | 20/238 [00:00<00:01, 192.41iter/s]
Build 3D deformation field: 17%|█████▎ | 41/238 [00:00<00:00, 199.91iter/s]
Build 3D deformation field: 26%|████████ | 62/238 [00:00<00:00, 200.76iter/s]
Build 3D deformation field: 35%|██████████▊ | 83/238 [00:00<00:00, 200.99iter/s]
Build 3D deformation field: 44%|█████████████ | 104/238 [00:00<00:00, 201.44iter/s]
Build 3D deformation field: 53%|███████████████▊ | 125/238 [00:00<00:00, 203.93iter/s]
Build 3D deformation field: 62%|██████████████████▌ | 147/238 [00:00<00:00, 206.30iter/s]
Build 3D deformation field: 71%|█████████████████████▏ | 168/238 [00:00<00:00, 204.54iter/s]
Build 3D deformation field: 79%|███████████████████████▊ | 189/238 [00:00<00:00, 203.30iter/s]
Build 3D deformation field: 88%|██████████████████████████▍ | 210/238 [00:01<00:00, 202.40iter/s]
Build 3D deformation field: 97%|█████████████████████████████ | 231/238 [00:01<00:00, 201.40iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 238/238 [00:01<00:00, 202.32iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
rm -rf /tmp/sct_2025-03-20_02-52-36_register-slicewise_uyymd0dk
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90/warp_src2dest.nii.gz warp_sub-NTNU1_acq-coilQaSagLarge_SNR_T00002sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU1_acq-coilQaSagLarge_SNR_T00002sub-NTNU1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-NTNU1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90/warp_dest2src.nii.gz warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-35_register-wrapper_owln7f90
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025240.990747.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-NTNU2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 71, 234)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-44_register-slicewise_41cunaa2)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 71 x 234
voxel size: 0.7065217mm x 0.6999992mm x 234mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/234 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/234 [00:00<00:50, 4.64iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
Estimate cord angle for each slice: 52%|███████████▍ | 122/234 [00:00<00:00, 484.41iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #216 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #217 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #218 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #219 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #220 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #223 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #224 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #225 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #226 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #227 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #228 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #229 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #230 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #231 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #232 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #233 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 234/234 [00:00<00:00, 583.94iter/s]
Build 3D deformation field: 0%| | 0/211 [00:00<?, ?iter/s]
Build 3D deformation field: 8%|██▍ | 17/211 [00:00<00:01, 162.50iter/s]
Build 3D deformation field: 17%|█████▏ | 35/211 [00:00<00:01, 168.56iter/s]
Build 3D deformation field: 25%|███████▊ | 53/211 [00:00<00:00, 170.25iter/s]
Build 3D deformation field: 34%|██████████▍ | 71/211 [00:00<00:00, 171.22iter/s]
Build 3D deformation field: 42%|█████████████ | 89/211 [00:00<00:00, 171.86iter/s]
Build 3D deformation field: 51%|███████████████▏ | 107/211 [00:00<00:00, 174.44iter/s]
Build 3D deformation field: 59%|█████████████████▊ | 125/211 [00:00<00:00, 174.51iter/s]
Build 3D deformation field: 68%|████████████████████▎ | 143/211 [00:00<00:00, 174.91iter/s]
Build 3D deformation field: 76%|██████████████████████▉ | 161/211 [00:00<00:00, 176.22iter/s]
Build 3D deformation field: 85%|█████████████████████████▍ | 179/211 [00:01<00:00, 175.93iter/s]
Build 3D deformation field: 93%|████████████████████████████ | 197/211 [00:01<00:00, 174.87iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 211/211 [00:01<00:00, 173.77iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
rm -rf /tmp/sct_2025-03-20_02-52-44_register-slicewise_41cunaa2
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64/warp_src2dest.nii.gz warp_sub-NTNU2_acq-coilQaSagLarge_SNR_T00002sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU2_acq-coilQaSagLarge_SNR_T00002sub-NTNU2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-NTNU2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64/warp_dest2src.nii.gz warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-43_register-wrapper_ssx9gz64
Finished! Elapsed time: 6s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025248.986365.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-NTNU3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz (60, 103, 322)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-52-53_register-slicewise_nmpgk2lu)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 60 x 103 x 322
voxel size: 0.7065217mm x 0.6999952mm x 322mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/322 [00:00<?, ?iter/s]
Estimate cord angle for each slice: 0%| | 1/322 [00:00<01:09, 4.62iter/s]
Estimate cord angle for each slice: 38%|████████▎ | 122/322 [00:00<00:00, 483.93iter/s]
Estimate cord angle for each slice: 76%|████████████████▋ | 244/322 [00:00<00:00, 749.53iter/s]
Estimate cord angle for each slice: 100%|██████████████████████| 322/322 [00:00<00:00, 669.31iter/s]
Build 3D deformation field: 0%| | 0/322 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 14/322 [00:00<00:02, 132.60iter/s]
Build 3D deformation field: 9%|██▊ | 29/322 [00:00<00:02, 138.04iter/s]
Build 3D deformation field: 14%|████▏ | 44/322 [00:00<00:01, 140.46iter/s]
Build 3D deformation field: 18%|█████▋ | 59/322 [00:00<00:01, 141.35iter/s]
Build 3D deformation field: 23%|███████ | 74/322 [00:00<00:01, 143.25iter/s]
Build 3D deformation field: 28%|████████▌ | 89/322 [00:00<00:01, 144.50iter/s]
Build 3D deformation field: 32%|█████████▋ | 104/322 [00:00<00:01, 144.68iter/s]
Build 3D deformation field: 37%|███████████ | 119/322 [00:00<00:01, 142.86iter/s]
Build 3D deformation field: 42%|████████████▍ | 134/322 [00:00<00:01, 143.47iter/s]
Build 3D deformation field: 46%|█████████████▉ | 149/322 [00:01<00:01, 144.64iter/s]
Build 3D deformation field: 51%|███████████████▎ | 164/322 [00:01<00:01, 144.95iter/s]
Build 3D deformation field: 56%|████████████████▋ | 179/322 [00:01<00:00, 144.39iter/s]
Build 3D deformation field: 60%|██████████████████ | 194/322 [00:01<00:00, 144.50iter/s]
Build 3D deformation field: 65%|███████████████████▍ | 209/322 [00:01<00:00, 145.37iter/s]
Build 3D deformation field: 70%|████████████████████▊ | 224/322 [00:01<00:00, 145.49iter/s]
Build 3D deformation field: 74%|██████████████████████▎ | 239/322 [00:01<00:00, 145.53iter/s]
Build 3D deformation field: 79%|███████████████████████▋ | 254/322 [00:01<00:00, 143.74iter/s]
Build 3D deformation field: 84%|█████████████████████████ | 269/322 [00:01<00:00, 144.14iter/s]
Build 3D deformation field: 88%|██████████████████████████▍ | 284/322 [00:01<00:00, 144.10iter/s]
Build 3D deformation field: 93%|███████████████████████████▊ | 299/322 [00:02<00:00, 144.00iter/s]
Build 3D deformation field: 98%|█████████████████████████████▎| 314/322 [00:02<00:00, 143.92iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 322/322 [00:02<00:00, 143.71iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
rm -rf /tmp/sct_2025-03-20_02-52-53_register-slicewise_nmpgk2lu
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6/warp_src2dest.nii.gz warp_sub-NTNU3_acq-coilQaSagLarge_SNR_T00002sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-NTNU3_acq-coilQaSagLarge_SNR_T00002sub-NTNU3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-NTNU3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6/warp_dest2src.nii.gz warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-52-51_register-wrapper_3mdvkvt6
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Converting image from type 'uint8' to type 'float64' for linear interpolation
Find the center of each slice
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025300.226075.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-UCL1_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz (67, 105, 333)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-53-05_register-slicewise_v2ase47u)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 67 x 105 x 333
voxel size: 0.7065217mm x 0.7mm x 333mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/333 [00:00<?, ?iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/333 [00:00<01:11, 4.63iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #6 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #7 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #8 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #9 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #10 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #11 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #12 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #13 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #14 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #15 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #16 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #17 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #18 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #19 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #20 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #21 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #22 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #23 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #24 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #25 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #26 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #27 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #28 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #29 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #30 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #31 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #32 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #33 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #34 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #35 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #36 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #37 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #38 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #39 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #40 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #41 is empty. It will be ignored.
Estimate cord angle for each slice: 42%|█████████▏ | 140/333 [00:00<00:00, 555.98iter/s]
Estimate cord angle for each slice: 79%|█████████████████▎ | 262/333 [00:00<00:00, 794.78iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #324 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #325 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #326 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #327 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #328 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #329 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #330 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #331 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #332 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 333/333 [00:00<00:00, 703.71iter/s]
Build 3D deformation field: 0%| | 0/282 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 12/282 [00:00<00:02, 112.02iter/s]
Build 3D deformation field: 9%|██▋ | 25/282 [00:00<00:02, 117.57iter/s]
Build 3D deformation field: 13%|████▏ | 38/282 [00:00<00:02, 119.84iter/s]
Build 3D deformation field: 18%|█████▌ | 51/282 [00:00<00:01, 120.83iter/s]
Build 3D deformation field: 23%|███████ | 64/282 [00:00<00:01, 121.79iter/s]
Build 3D deformation field: 27%|████████▍ | 77/282 [00:00<00:01, 122.30iter/s]
Build 3D deformation field: 32%|█████████▉ | 90/282 [00:00<00:01, 124.11iter/s]
Build 3D deformation field: 37%|██████████▉ | 103/282 [00:00<00:01, 124.28iter/s]
Build 3D deformation field: 41%|████████████▎ | 116/282 [00:00<00:01, 124.76iter/s]
Build 3D deformation field: 46%|█████████████▋ | 129/282 [00:01<00:01, 125.17iter/s]
Build 3D deformation field: 50%|███████████████ | 142/282 [00:01<00:01, 126.16iter/s]
Build 3D deformation field: 55%|████████████████▍ | 155/282 [00:01<00:01, 126.88iter/s]
Build 3D deformation field: 60%|█████████████████▊ | 168/282 [00:01<00:00, 127.36iter/s]
Build 3D deformation field: 64%|███████████████████▎ | 181/282 [00:01<00:00, 127.74iter/s]
Build 3D deformation field: 69%|████████████████████▋ | 194/282 [00:01<00:00, 127.17iter/s]
Build 3D deformation field: 73%|██████████████████████ | 207/282 [00:01<00:00, 126.26iter/s]
Build 3D deformation field: 78%|███████████████████████▍ | 220/282 [00:01<00:00, 126.36iter/s]
Build 3D deformation field: 83%|████████████████████████▊ | 233/282 [00:01<00:00, 126.83iter/s]
Build 3D deformation field: 87%|██████████████████████████▏ | 246/282 [00:01<00:00, 126.75iter/s]
Build 3D deformation field: 92%|███████████████████████████▌ | 259/282 [00:02<00:00, 127.20iter/s]
Build 3D deformation field: 96%|████████████████████████████▉ | 272/282 [00:02<00:00, 127.10iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 282/282 [00:02<00:00, 125.28iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
rm -rf /tmp/sct_2025-03-20_02-53-05_register-slicewise_v2ase47u
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL1_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v/warp_src2dest.nii.gz warp_sub-UCL1_acq-coilQaSagLarge_SNR_T00002sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL1_acq-coilQaSagLarge_SNR_T00002sub-UCL1_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-UCL1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-UCL1_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v/warp_dest2src.nii.gz warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-53-03_register-wrapper_ge58xd6v
Finished! Elapsed time: 10s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025312.667511.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-UCL2_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz (75, 110, 287)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-53-18_register-slicewise_v7ecmhu3)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 75 x 110 x 287
voxel size: 0.7065217mm x 0.7000031mm x 287mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/287 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/287 [00:00<01:03, 4.52iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 41%|█████████ | 119/287 [00:00<00:00, 464.66iter/s]
Estimate cord angle for each slice: 83%|██████████████████▎ | 239/287 [00:00<00:00, 727.29iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #258 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #259 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #260 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #261 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #262 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #263 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #264 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #265 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #266 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #267 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #268 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #269 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 287/287 [00:00<00:00, 630.83iter/s]
Build 3D deformation field: 0%| | 0/255 [00:00<?, ?iter/s]
Build 3D deformation field: 4%|█▎ | 11/255 [00:00<00:02, 104.01iter/s]
Build 3D deformation field: 9%|██▋ | 22/255 [00:00<00:02, 106.41iter/s]
Build 3D deformation field: 13%|████ | 33/255 [00:00<00:02, 106.24iter/s]
Build 3D deformation field: 17%|█████▎ | 44/255 [00:00<00:01, 107.28iter/s]
Build 3D deformation field: 22%|██████▋ | 55/255 [00:00<00:01, 107.32iter/s]
Build 3D deformation field: 26%|████████ | 66/255 [00:00<00:01, 107.89iter/s]
Build 3D deformation field: 30%|█████████▎ | 77/255 [00:00<00:01, 108.21iter/s]
Build 3D deformation field: 35%|██████████▋ | 88/255 [00:00<00:01, 107.20iter/s]
Build 3D deformation field: 39%|████████████ | 99/255 [00:00<00:01, 105.97iter/s]
Build 3D deformation field: 43%|████████████▉ | 110/255 [00:01<00:01, 105.24iter/s]
Build 3D deformation field: 47%|██████████████▏ | 121/255 [00:01<00:01, 106.60iter/s]
Build 3D deformation field: 52%|███████████████▌ | 132/255 [00:01<00:01, 107.45iter/s]
Build 3D deformation field: 56%|████████████████▊ | 143/255 [00:01<00:01, 107.81iter/s]
Build 3D deformation field: 60%|██████████████████ | 154/255 [00:01<00:00, 108.18iter/s]
Build 3D deformation field: 65%|███████████████████▍ | 165/255 [00:01<00:00, 107.80iter/s]
Build 3D deformation field: 69%|████████████████████▋ | 176/255 [00:01<00:00, 108.22iter/s]
Build 3D deformation field: 73%|██████████████████████ | 187/255 [00:01<00:00, 108.43iter/s]
Build 3D deformation field: 78%|███████████████████████▎ | 198/255 [00:01<00:00, 108.85iter/s]
Build 3D deformation field: 82%|████████████████████████▌ | 209/255 [00:01<00:00, 108.62iter/s]
Build 3D deformation field: 86%|█████████████████████████▉ | 220/255 [00:02<00:00, 108.94iter/s]
Build 3D deformation field: 91%|███████████████████████████▏ | 231/255 [00:02<00:00, 108.93iter/s]
Build 3D deformation field: 95%|████████████████████████████▍ | 242/255 [00:02<00:00, 108.51iter/s]
Build 3D deformation field: 99%|█████████████████████████████▊| 253/255 [00:02<00:00, 108.13iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 255/255 [00:02<00:00, 107.69iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
rm -rf /tmp/sct_2025-03-20_02-53-18_register-slicewise_v7ecmhu3
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL2_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8/warp_src2dest.nii.gz warp_sub-UCL2_acq-coilQaSagLarge_SNR_T00002sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL2_acq-coilQaSagLarge_SNR_T00002sub-UCL2_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-UCL2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-UCL2_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8/warp_dest2src.nii.gz warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-53-15_register-wrapper_57rk07e8
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025325.189883.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -d ../anat/sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz -iseg sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dseg ../anat/sub-UCL3_UNIT1_seg_crop.nii.gz -param step=1,type=seg,algo=centermass -qc /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc
--
Input parameters:
Source .............. sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../anat/sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz (69, 115, 300)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... centermass
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Creating temporary folder (/tmp/sct_2025-03-20_02-53-30_register-slicewise_n1di0jde)
Copy input data to temp folder...
Get image dimensions of destination image...
matrix size: 69 x 115 x 300
voxel size: 0.7065217mm x 0.7mm x 300mm
Split input segmentation...
Split destination segmentation...
Estimate cord angle for each slice: 0%| | 0/300 [00:00<?, ?iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #0 is empty. It will be ignored.
Estimate cord angle for each slice: 0%| | 1/300 [00:00<01:05, 4.53iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #1 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #2 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #3 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #4 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #5 is empty. It will be ignored.
Estimate cord angle for each slice: 40%|████████▋ | 119/300 [00:00<00:00, 465.41iter/s]
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #180 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #181 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #182 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #183 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #216 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #217 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #218 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #219 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #220 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #221 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #222 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #223 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #224 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #225 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #226 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #227 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #228 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #229 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #230 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #231 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #232 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #233 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #234 is empty. It will be ignored.
Estimate cord angle for each slice: 82%|█████████████████▉ | 245/300 [00:00<00:00, 748.58iter/s]/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #252 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #253 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #254 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #255 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #256 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #257 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #258 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #259 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #260 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #261 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #262 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #263 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #264 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #265 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #266 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #267 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #268 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #269 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #270 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #271 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #272 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #273 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #274 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #275 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #276 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #277 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #278 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #279 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #280 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #281 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #282 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #283 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #284 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #285 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #286 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #287 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #288 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #289 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #290 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #291 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #292 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #293 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #294 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #295 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #296 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #297 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #298 is empty. It will be ignored.
/home/runner/sct_6.5/spinalcordtoolbox/registration/algorithms.py:1440: RuntimeWarning: Mean of empty slice.
centermass = coordsrc.mean(0)
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:121: RuntimeWarning: invalid value encountered in divide
ret = um.true_divide(
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:206: RuntimeWarning: Degrees of freedom <= 0 for slice
ret = _var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:163: RuntimeWarning: invalid value encountered in divide
arrmean = um.true_divide(arrmean, div, out=arrmean,
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/numpy/core/_methods.py:198: RuntimeWarning: invalid value encountered in scalar divide
ret = ret.dtype.type(ret / rcount)
Slice #299 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|██████████████████████| 300/300 [00:00<00:00, 663.76iter/s]
Build 3D deformation field: 0%| | 0/223 [00:00<?, ?iter/s]
Build 3D deformation field: 5%|█▌ | 11/223 [00:00<00:02, 103.04iter/s]
Build 3D deformation field: 10%|███ | 22/223 [00:00<00:01, 105.37iter/s]
Build 3D deformation field: 15%|████▌ | 33/223 [00:00<00:01, 106.26iter/s]
Build 3D deformation field: 20%|██████▎ | 45/223 [00:00<00:01, 108.24iter/s]
Build 3D deformation field: 26%|███████▉ | 57/223 [00:00<00:01, 109.86iter/s]
Build 3D deformation field: 31%|█████████▌ | 69/223 [00:00<00:01, 110.92iter/s]
Build 3D deformation field: 36%|███████████▎ | 81/223 [00:00<00:01, 111.58iter/s]
Build 3D deformation field: 42%|████████████▉ | 93/223 [00:00<00:01, 112.02iter/s]
Build 3D deformation field: 47%|██████████████▏ | 105/223 [00:00<00:01, 112.35iter/s]
Build 3D deformation field: 52%|███████████████▋ | 117/223 [00:01<00:00, 112.27iter/s]
Build 3D deformation field: 58%|█████████████████▎ | 129/223 [00:01<00:00, 111.50iter/s]
Build 3D deformation field: 63%|██████████████████▉ | 141/223 [00:01<00:00, 111.89iter/s]
Build 3D deformation field: 69%|████████████████████▌ | 153/223 [00:01<00:00, 111.96iter/s]
Build 3D deformation field: 74%|██████████████████████▏ | 165/223 [00:01<00:00, 111.86iter/s]
Build 3D deformation field: 79%|███████████████████████▊ | 177/223 [00:01<00:00, 111.51iter/s]
Build 3D deformation field: 85%|█████████████████████████▍ | 189/223 [00:01<00:00, 111.89iter/s]
Build 3D deformation field: 90%|███████████████████████████ | 201/223 [00:01<00:00, 112.10iter/s]
Build 3D deformation field: 96%|████████████████████████████▋ | 213/223 [00:01<00:00, 111.09iter/s]
Build 3D deformation field: 100%|██████████████████████████████| 223/223 [00:02<00:00, 110.99iter/s]
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1Warp.nii.gz
Generate warping field...
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--> step1InverseWarp.nii.gz
Move warping fields...
cp step1Warp.nii.gz /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
cp step1InverseWarp.nii.gz /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
rm -rf /tmp/sct_2025-03-20_02-53-30_register-slicewise_n1di0jde
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii warp_inverse_0.nii.gz warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp/warp_src2dest.nii.gz warp_sub-UCL3_acq-coilQaSagLarge_SNR_T00002sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File created: warp_sub-UCL3_acq-coilQaSagLarge_SNR_T00002sub-UCL3_inv-1_part-mag_MP2RAGE_crop.nii.gz
File sub-UCL3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz already exists. Deleting it..
File created: sub-UCL3_inv-1_part-mag_MP2RAGE_crop_reg.nii.gz
mv /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp/warp_dest2src.nii.gz warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-53-28_register-wrapper_m4utn4vp
Finished! Elapsed time: 9s
*** Generating Quality Control (QC) html report ***
Resample images to 0.600000x0.600000 mm
Converting image from type 'int16' to type 'float64' for spline interpolation
Find the center of each slice
/home/runner/sct_6.5/python/envs/venv_sct/lib/python3.9/site-packages/scipy/ndimage/_measurements.py:1542: RuntimeWarning: invalid value encountered in scalar divide
results = [sum(input * grids[dir].astype(float), labels, index) / normalizer
Successfully generated the QC results in /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/_json/qc_2025_03_20_025337.449819.json
To see the results in a browser, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/qc/index.html
Warp MP2RAGE vertebral levels to fmap metrics#
# Warping vertebral level to each flip angle and SNR map
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
# For MSSM subjects, we also need to warp the SC segmentation
if subject in ['sub-MSSM1', 'sub-MSSM2', 'sub-MSSM3']:
# Warping spinal cord segmentation to TFL data
!sct_apply_transfo -i ../anat/{subject}_UNIT1_seg_crop.nii.gz -d {subject}_acq-famp_TB1TFL.nii.gz -w warp_{subject}_inv-1_part-mag_MP2RAGE_crop2{subject}_acq-famp_TB1TFL.nii.gz -x linear -o {subject}_acq-anat_TB1TFL_seg.nii.gz
# Setting type to 'famp' for command below (https://github.com/spinal-cord-7t/coil-qc-code/issues/43)
type = 'famp'
else:
type = 'anat'
# Warping vertebral levels to TFL data
!sct_apply_transfo -i ../anat/{subject}_UNIT1_seg_labeled_crop.nii.gz -d {subject}_acq-famp_TB1TFL.nii.gz -w warp_{subject}_inv-1_part-mag_MP2RAGE_crop2{subject}_acq-{type}_TB1TFL.nii.gz -x nn -o {subject}_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
# !sct_qc -i {subject}_acq-{type}_TB1TFL.nii.gz -s {subject}_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -p sct_label_vertebrae -qc {path_qc} -qc-subject {subject}
# Warping SC segmentation and vertebral levels to DREAM fmaps
!sct_apply_transfo -i ../anat/{subject}_UNIT1_seg_crop.nii.gz -d {subject}_acq-famp_TB1DREAM.nii.gz -w warp_{subject}_UNIT1_crop2{subject}_acq-famp_TB1DREAM.nii.gz -x linear -o {subject}_acq-famp_TB1DREAM_seg.nii.gz
# !sct_qc -i {subject}_acq-famp_TB1DREAM.nii.gz -s {subject}_acq-famp_TB1DREAM_seg.nii.gz -p sct_deepseg_sc -qc {path_qc} -qc-subject {subject}
!sct_apply_transfo -i ../anat/{subject}_UNIT1_seg_labeled_crop.nii.gz -d {subject}_acq-famp_TB1DREAM.nii.gz -w warp_{subject}_UNIT1_crop2{subject}_acq-famp_TB1DREAM.nii.gz -x nn -o {subject}_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
# !sct_qc -i {subject}_acq-famp_TB1DREAM.nii.gz -s {subject}_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -p sct_label_vertebrae -qc {path_qc} -qc-subject {subject}
# Warping SC segmentation and vertebral level to SNR maps
!sct_apply_transfo -i ../anat/{subject}_UNIT1_seg_labeled_crop.nii.gz -d {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_{subject}_inv-1_part-mag_MP2RAGE_crop2{subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o {subject}_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
# !sct_qc -i {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -s {subject}_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -p sct_label_vertebrae -qc {path_qc} -qc-subject {subject}
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM1_acq-famp_TB1TFL.nii.gz -w warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-anat_TB1TFL.nii.gz -x nn -o sub-CRMBM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 100 x 314 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-anat_TB1TFL.nii.gz -r sub-CRMBM1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM1_UNIT1_seg_crop.nii.gz -d sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-CRMBM1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 100 x 314 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM1_UNIT1_seg_crop.nii.gz -o sub-CRMBM1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-CRMBM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 100 x 314 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM1_UNIT1_crop2sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 100 x 314 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM1_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM1_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM2_acq-famp_TB1TFL.nii.gz -w warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-anat_TB1TFL.nii.gz -x nn -o sub-CRMBM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 90 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-anat_TB1TFL.nii.gz -r sub-CRMBM2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM2_UNIT1_seg_crop.nii.gz -d sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-CRMBM2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 90 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM2_UNIT1_seg_crop.nii.gz -o sub-CRMBM2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-CRMBM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 90 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM2_UNIT1_crop2sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 90 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM2_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM2_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM3_acq-famp_TB1TFL.nii.gz -w warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-anat_TB1TFL.nii.gz -x nn -o sub-CRMBM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 121 x 337 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-anat_TB1TFL.nii.gz -r sub-CRMBM3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM3_UNIT1_seg_crop.nii.gz -d sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 121 x 337 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM3_UNIT1_seg_crop.nii.gz -o sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -w warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-CRMBM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 121 x 337 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM3_UNIT1_crop2sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -r sub-CRMBM3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -d sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 121 x 337 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-CRMBM3_UNIT1_seg_labeled_crop.nii.gz -o sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-CRMBM3_inv-1_part-mag_MP2RAGE_crop2sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-CRMBM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH1_acq-famp_TB1TFL.nii.gz -w warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-anat_TB1TFL.nii.gz -x nn -o sub-MGH1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 118 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-anat_TB1TFL.nii.gz -r sub-MGH1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH1_UNIT1_seg_crop.nii.gz -d sub-MGH1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MGH1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 118 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH1_UNIT1_seg_crop.nii.gz -o sub-MGH1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz -r sub-MGH1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MGH1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 118 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH1_UNIT1_crop2sub-MGH1_acq-famp_TB1DREAM.nii.gz -r sub-MGH1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 118 x 226 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH1_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH1_inv-1_part-mag_MP2RAGE_crop2sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH2_acq-famp_TB1TFL.nii.gz -w warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-anat_TB1TFL.nii.gz -x nn -o sub-MGH2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
59 x 117 x 223 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-anat_TB1TFL.nii.gz -r sub-MGH2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH2_UNIT1_seg_crop.nii.gz -d sub-MGH2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MGH2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
59 x 117 x 223 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH2_UNIT1_seg_crop.nii.gz -o sub-MGH2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz -r sub-MGH2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MGH2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
59 x 117 x 223 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH2_UNIT1_crop2sub-MGH2_acq-famp_TB1DREAM.nii.gz -r sub-MGH2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
59 x 117 x 223 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH2_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH2_inv-1_part-mag_MP2RAGE_crop2sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH3_acq-famp_TB1TFL.nii.gz -w warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-anat_TB1TFL.nii.gz -x nn -o sub-MGH3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 127 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-anat_TB1TFL.nii.gz -r sub-MGH3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH3_UNIT1_seg_crop.nii.gz -d sub-MGH3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MGH3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 127 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH3_UNIT1_seg_crop.nii.gz -o sub-MGH3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz -r sub-MGH3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MGH3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 127 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH3_UNIT1_crop2sub-MGH3_acq-famp_TB1DREAM.nii.gz -r sub-MGH3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -d sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 127 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MGH3_UNIT1_seg_labeled_crop.nii.gz -o sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MGH3_inv-1_part-mag_MP2RAGE_crop2sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MGH3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI1_acq-famp_TB1TFL.nii.gz -w warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-anat_TB1TFL.nii.gz -x nn -o sub-MNI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 77 x 367 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-anat_TB1TFL.nii.gz -r sub-MNI1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI1_UNIT1_seg_crop.nii.gz -d sub-MNI1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MNI1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 77 x 367 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI1_UNIT1_seg_crop.nii.gz -o sub-MNI1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz -r sub-MNI1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MNI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 77 x 367 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI1_UNIT1_crop2sub-MNI1_acq-famp_TB1DREAM.nii.gz -r sub-MNI1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 77 x 367 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI1_inv-1_part-mag_MP2RAGE_crop2sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI2_acq-famp_TB1TFL.nii.gz -w warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-anat_TB1TFL.nii.gz -x nn -o sub-MNI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 75 x 361 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-anat_TB1TFL.nii.gz -r sub-MNI2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI2_UNIT1_seg_crop.nii.gz -d sub-MNI2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MNI2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 75 x 361 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI2_UNIT1_seg_crop.nii.gz -o sub-MNI2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz -r sub-MNI2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MNI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 75 x 361 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI2_UNIT1_crop2sub-MNI2_acq-famp_TB1DREAM.nii.gz -r sub-MNI2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
63 x 75 x 361 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI2_inv-1_part-mag_MP2RAGE_crop2sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI3_acq-famp_TB1TFL.nii.gz -w warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-anat_TB1TFL.nii.gz -x nn -o sub-MNI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 102 x 260 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-anat_TB1TFL.nii.gz -r sub-MNI3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI3_UNIT1_seg_crop.nii.gz -d sub-MNI3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MNI3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 102 x 260 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI3_UNIT1_seg_crop.nii.gz -o sub-MNI3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz -r sub-MNI3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MNI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 102 x 260 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI3_UNIT1_crop2sub-MNI3_acq-famp_TB1DREAM.nii.gz -r sub-MNI3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 102 x 260 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MNI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MNI3_inv-1_part-mag_MP2RAGE_crop2sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MNI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI1_acq-famp_TB1TFL.nii.gz -w warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-anat_TB1TFL.nii.gz -x nn -o sub-MPI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 89 x 266 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-anat_TB1TFL.nii.gz -r sub-MPI1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI1_UNIT1_seg_crop.nii.gz -d sub-MPI1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MPI1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 89 x 266 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI1_UNIT1_seg_crop.nii.gz -o sub-MPI1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz -r sub-MPI1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MPI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 89 x 266 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI1_UNIT1_crop2sub-MPI1_acq-famp_TB1DREAM.nii.gz -r sub-MPI1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 89 x 266 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI1_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI1_inv-1_part-mag_MP2RAGE_crop2sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI2_acq-famp_TB1TFL.nii.gz -w warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-anat_TB1TFL.nii.gz -x nn -o sub-MPI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
61 x 89 x 276 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-anat_TB1TFL.nii.gz -r sub-MPI2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI2_UNIT1_seg_crop.nii.gz -d sub-MPI2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MPI2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
61 x 89 x 276 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI2_UNIT1_seg_crop.nii.gz -o sub-MPI2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz -r sub-MPI2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MPI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
61 x 89 x 276 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI2_UNIT1_crop2sub-MPI2_acq-famp_TB1DREAM.nii.gz -r sub-MPI2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
61 x 89 x 276 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI2_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI2_inv-1_part-mag_MP2RAGE_crop2sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI3_acq-famp_TB1TFL.nii.gz -w warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-anat_TB1TFL.nii.gz -x nn -o sub-MPI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 117 x 282 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-anat_TB1TFL.nii.gz -r sub-MPI3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI3_UNIT1_seg_crop.nii.gz -d sub-MPI3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MPI3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 117 x 282 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI3_UNIT1_seg_crop.nii.gz -o sub-MPI3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz -r sub-MPI3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MPI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 117 x 282 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI3_UNIT1_crop2sub-MPI3_acq-famp_TB1DREAM.nii.gz -r sub-MPI3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -d sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 117 x 282 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MPI3_UNIT1_seg_labeled_crop.nii.gz -o sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MPI3_inv-1_part-mag_MP2RAGE_crop2sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MPI3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -d sub-MSSM1_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz -x linear -o sub-MSSM1_acq-anat_TB1TFL_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 72 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -o sub-MSSM1_acq-anat_TB1TFL_seg.nii.gz -t warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz -r sub-MSSM1_acq-famp_TB1TFL.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM1_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz -x nn -o sub-MSSM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 72 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-famp_TB1TFL.nii.gz -r sub-MSSM1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -d sub-MSSM1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MSSM1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 72 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM1_UNIT1_seg_crop.nii.gz -o sub-MSSM1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz -r sub-MSSM1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM1_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MSSM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 72 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM1_UNIT1_crop2sub-MSSM1_acq-famp_TB1DREAM.nii.gz -r sub-MSSM1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
68 x 72 x 238 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM1_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM1_inv-1_part-mag_MP2RAGE_crop2sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -d sub-MSSM2_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz -x linear -o sub-MSSM2_acq-anat_TB1TFL_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 74 x 228 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -o sub-MSSM2_acq-anat_TB1TFL_seg.nii.gz -t warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz -r sub-MSSM2_acq-famp_TB1TFL.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM2_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz -x nn -o sub-MSSM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 74 x 228 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-famp_TB1TFL.nii.gz -r sub-MSSM2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -d sub-MSSM2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MSSM2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 74 x 228 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM2_UNIT1_seg_crop.nii.gz -o sub-MSSM2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz -r sub-MSSM2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM2_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MSSM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 74 x 228 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM2_UNIT1_crop2sub-MSSM2_acq-famp_TB1DREAM.nii.gz -r sub-MSSM2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
62 x 74 x 228 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM2_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM2_inv-1_part-mag_MP2RAGE_crop2sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -d sub-MSSM3_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz -x linear -o sub-MSSM3_acq-anat_TB1TFL_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 104 x 289 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -o sub-MSSM3_acq-anat_TB1TFL_seg.nii.gz -t warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz -r sub-MSSM3_acq-famp_TB1TFL.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM3_acq-famp_TB1TFL.nii.gz -w warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz -x nn -o sub-MSSM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 104 x 289 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-famp_TB1TFL.nii.gz -r sub-MSSM3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -d sub-MSSM3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-MSSM3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 104 x 289 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM3_UNIT1_seg_crop.nii.gz -o sub-MSSM3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz -r sub-MSSM3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM3_acq-famp_TB1DREAM.nii.gz -w warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-MSSM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 104 x 289 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM3_UNIT1_crop2sub-MSSM3_acq-famp_TB1DREAM.nii.gz -r sub-MSSM3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -d sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 104 x 289 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-MSSM3_UNIT1_seg_labeled_crop.nii.gz -o sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-MSSM3_inv-1_part-mag_MP2RAGE_crop2sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-MSSM3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU1_acq-famp_TB1TFL.nii.gz -w warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-anat_TB1TFL.nii.gz -x nn -o sub-NTNU1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 65 x 296 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-anat_TB1TFL.nii.gz -r sub-NTNU1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU1_UNIT1_seg_crop.nii.gz -d sub-NTNU1_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-NTNU1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 65 x 296 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU1_UNIT1_seg_crop.nii.gz -o sub-NTNU1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz -r sub-NTNU1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU1_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-NTNU1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 65 x 296 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU1_UNIT1_crop2sub-NTNU1_acq-famp_TB1DREAM.nii.gz -r sub-NTNU1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
65 x 65 x 296 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU1_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU1_inv-1_part-mag_MP2RAGE_crop2sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU2_acq-famp_TB1TFL.nii.gz -w warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-anat_TB1TFL.nii.gz -x nn -o sub-NTNU2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 71 x 234 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-anat_TB1TFL.nii.gz -r sub-NTNU2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU2_UNIT1_seg_crop.nii.gz -d sub-NTNU2_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-NTNU2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 71 x 234 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU2_UNIT1_seg_crop.nii.gz -o sub-NTNU2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz -r sub-NTNU2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU2_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-NTNU2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 71 x 234 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU2_UNIT1_crop2sub-NTNU2_acq-famp_TB1DREAM.nii.gz -r sub-NTNU2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 71 x 234 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU2_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU2_inv-1_part-mag_MP2RAGE_crop2sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU3_acq-famp_TB1TFL.nii.gz -w warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-anat_TB1TFL.nii.gz -x nn -o sub-NTNU3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
60 x 103 x 322 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-anat_TB1TFL.nii.gz -r sub-NTNU3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU3_UNIT1_seg_crop.nii.gz -d sub-NTNU3_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-NTNU3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
60 x 103 x 322 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU3_UNIT1_seg_crop.nii.gz -o sub-NTNU3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz -r sub-NTNU3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU3_acq-famp_TB1DREAM.nii.gz -w warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-NTNU3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
60 x 103 x 322 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU3_UNIT1_crop2sub-NTNU3_acq-famp_TB1DREAM.nii.gz -r sub-NTNU3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -d sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
60 x 103 x 322 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-NTNU3_UNIT1_seg_labeled_crop.nii.gz -o sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-NTNU3_inv-1_part-mag_MP2RAGE_crop2sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-NTNU3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL1_acq-famp_TB1TFL.nii.gz -w warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-anat_TB1TFL.nii.gz -x nn -o sub-UCL1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
67 x 105 x 333 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-anat_TB1TFL.nii.gz -r sub-UCL1_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL1_UNIT1_seg_crop.nii.gz -d sub-UCL1_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz -x linear -o sub-UCL1_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
67 x 105 x 333 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL1_UNIT1_seg_crop.nii.gz -o sub-UCL1_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz -r sub-UCL1_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL1_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz -x nn -o sub-UCL1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
67 x 105 x 333 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL1_UNIT1_crop2sub-UCL1_acq-famp_TB1DREAM.nii.gz -r sub-UCL1_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
67 x 105 x 333 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL1_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL1_inv-1_part-mag_MP2RAGE_crop2sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL1/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL2_acq-famp_TB1TFL.nii.gz -w warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-anat_TB1TFL.nii.gz -x nn -o sub-UCL2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
75 x 110 x 287 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-anat_TB1TFL.nii.gz -r sub-UCL2_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL2_UNIT1_seg_crop.nii.gz -d sub-UCL2_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz -x linear -o sub-UCL2_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
75 x 110 x 287 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL2_UNIT1_seg_crop.nii.gz -o sub-UCL2_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz -r sub-UCL2_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL2_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz -x nn -o sub-UCL2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
75 x 110 x 287 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL2_UNIT1_crop2sub-UCL2_acq-famp_TB1DREAM.nii.gz -r sub-UCL2_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
75 x 110 x 287 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL2_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL2_inv-1_part-mag_MP2RAGE_crop2sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL2/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL3_acq-famp_TB1TFL.nii.gz -w warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-anat_TB1TFL.nii.gz -x nn -o sub-UCL3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 115 x 300 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-anat_TB1TFL.nii.gz -r sub-UCL3_acq-famp_TB1TFL.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL3_UNIT1_seg_crop.nii.gz -d sub-UCL3_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz -x linear -o sub-UCL3_acq-famp_TB1DREAM_seg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 115 x 300 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL3_UNIT1_seg_crop.nii.gz -o sub-UCL3_acq-famp_TB1DREAM_seg.nii.gz -t warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz -r sub-UCL3_acq-famp_TB1DREAM.nii.gz -n Linear # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL3_acq-famp_TB1DREAM.nii.gz -w warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz -x nn -o sub-UCL3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 115 x 300 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL3_UNIT1_crop2sub-UCL3_acq-famp_TB1DREAM.nii.gz -r sub-UCL3_acq-famp_TB1DREAM.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
--
Spinal Cord Toolbox (6.5)
sct_apply_transfo -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -d sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -w warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -x nn -o sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz
--
Parse list of warping fields...
Get dimensions of data...
69 x 115 x 300 x 1
Apply transformation...
Apply transformation and resample to destination space...
/home/runner/sct_6.5/bin/isct_antsApplyTransforms -d 3 -i ../anat/sub-UCL3_UNIT1_seg_labeled_crop.nii.gz -o sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -t warp_sub-UCL3_inv-1_part-mag_MP2RAGE_crop2sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -r sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -n NearestNeighbor # in /home/runner/work/coil-qc-code/coil-qc-code/data-human/sub-UCL3/fmap
Copy affine matrix from destination space to make sure qform/sform are the same.
Quality control (local station)#
# Quality control of registration
# This code generates syntax to open the registered data in FSLeyes. The syntax should be run from within the `data-human/` folder.
for subject in subjects:
# for subject in ['sub-MGH1']:
print(f"\n👉 CHECKING REGISTRATION FOR: {subject}\n")
cmd = f"fsleyes {subject}/fmap/{subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz {subject}/fmap/{subject}_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow"
print(cmd+" &")
# TODO: undisplay all scans but the first
👉 CHECKING REGISTRATION FOR: sub-CRMBM1
fsleyes sub-CRMBM1/fmap/sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-CRMBM1/fmap/sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-CRMBM2
fsleyes sub-CRMBM2/fmap/sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-CRMBM2/fmap/sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-CRMBM3
fsleyes sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MGH1
fsleyes sub-MGH1/fmap/sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MGH1/fmap/sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MGH2
fsleyes sub-MGH2/fmap/sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MGH2/fmap/sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MGH3
fsleyes sub-MGH3/fmap/sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MGH3/fmap/sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MNI1
fsleyes sub-MNI1/fmap/sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MNI1/fmap/sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MNI2
fsleyes sub-MNI2/fmap/sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MNI2/fmap/sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MNI3
fsleyes sub-MNI3/fmap/sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MNI3/fmap/sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MPI1
fsleyes sub-MPI1/fmap/sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MPI1/fmap/sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MPI2
fsleyes sub-MPI2/fmap/sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MPI2/fmap/sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MPI3
fsleyes sub-MPI3/fmap/sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MPI3/fmap/sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MSSM1
fsleyes sub-MSSM1/fmap/sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MSSM1/fmap/sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MSSM2
fsleyes sub-MSSM2/fmap/sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MSSM2/fmap/sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-MSSM3
fsleyes sub-MSSM3/fmap/sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-MSSM3/fmap/sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-NTNU1
fsleyes sub-NTNU1/fmap/sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-NTNU1/fmap/sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-NTNU2
fsleyes sub-NTNU2/fmap/sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-NTNU2/fmap/sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-NTNU3
fsleyes sub-NTNU3/fmap/sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-NTNU3/fmap/sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-UCL1
fsleyes sub-UCL1/fmap/sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-UCL1/fmap/sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-UCL2
fsleyes sub-UCL2/fmap/sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-UCL2/fmap/sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
👉 CHECKING REGISTRATION FOR: sub-UCL3
fsleyes sub-UCL3/fmap/sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz sub-UCL3/fmap/sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz --cmap rainbow &
Convert TFL and DREAM flip angle maps to B1+ in units of nT/V#
# load DREAM FA maps acquired with different reference voltages
# threshold FA maps to 20deg < FA < 50deg
# combine FA maps by averaging non-zero estimates of FA in each pixel
GAMMA = 2.675e8; # [rad / (s T)]
voltages = ["1.5", "0.66"]
for subject in subjects:
b1_maps = []
os.chdir(os.path.join(path_data, subject, "fmap"))
if subject=='sub-MSSM1':
ref_voltage=450
elif subject=='sub-MSSM2':
ref_voltage=350
elif subject=='sub-MSSM3':
ref_voltage=450
else:
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp_TB1DREAM.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Open refV flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp_TB1DREAM.nii.gz")
meas_fa = nii.get_fdata()
#thresholding
meas_fa[meas_fa < 200] = np.nan
meas_fa[meas_fa > 500] = np.nan
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp_TB1DREAM.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
#convert measured FA to percent of requested FA (note that measured FA map is in degrees * 10)
meas_fa = (meas_fa/10) / requested_fa
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
b1_map = meas_fa * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
b1_maps.append(b1_map)
for voltage in voltages:
#check if map exists
my_file = Path(f"{subject}_acq-famp-{voltage}_TB1DREAM.nii.gz")
if my_file.is_file():
if subject=='sub-MSSM2' and voltage=="1.5":
ref_voltage=450
elif subject=='sub-MSSM2' and voltage=="0.66":
ref_voltage=234
elif subject=='sub-MSSM3' and voltage=="0.66":
ref_voltage=328
else:
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp-{voltage}_TB1DREAM.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Open flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp-{voltage}_TB1DREAM.nii.gz")
meas_fa = nii.get_fdata()
#thresholding
meas_fa[meas_fa < 200] = np.nan
meas_fa[meas_fa > 500] = np.nan
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp-{voltage}_TB1DREAM.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
#convert measured FA to percent of requested FA (note that measured FA map is in degrees * 10)
meas_fa = (meas_fa/10) / requested_fa
else:
meas_fa = np.full((nii.header).get_data_shape(),np.nan)
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
# Siemens maps are in units of flip angle * 10 (in degrees)
b1_map = meas_fa * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
b1_maps.append(b1_map)
# compute mean of non-zero values
avgB1=np.nanmean(b1_maps,axis=0)
# Save as NIfTI file
nii_avgB1 = nib.Nifti1Image(avgB1, nii.affine, nii.header)
nib.save(nii_avgB1, f"{subject}_DREAMTB1avgB1map.nii.gz")
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
/tmp/ipykernel_2706/4246477433.py:104: RuntimeWarning: Mean of empty slice
avgB1=np.nanmean(b1_maps,axis=0)
# Convert the TFL flip angle maps to B1+ efficiency maps [nT/V] (inspired by code from Kyle Gilbert)
# The approach consists in calculating the B1+ efficiency using a 1ms, pi-pulse at the acquisition voltage,
# then scale the efficiency by the ratio of the measured flip angle to the requested flip angle in the pulse sequence.
GAMMA = 2.675e8; # [rad / (s T)]
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
if subject=='sub-MSSM1':
ref_voltage=450
elif subject=='sub-MSSM2':
ref_voltage=350
elif subject=='sub-MSSM3':
ref_voltage=450
else:
# Fetch the reference voltage from the JSON sidecar
with open(f"{subject}_acq-famp_TB1TFL.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
print(f"ref_voltage [V]: {ref_voltage} ({subject}_acq-famp_TB1TFL)")
# Fetch the flip angle from the JSON sidecar
with open(f"{subject}_acq-famp_TB1TFL.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
print(f"flip angle [degrees]: {requested_fa} ({subject}_acq-famp_TB1TFL)")
# Open flip angle map with nibabel
nii = nib.load(f"{subject}_acq-famp_TB1TFL.nii.gz")
meas_fa = nii.get_fdata()
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Compute B1 map in [T/V]
# Siemens maps are in units of flip angle * 10 (in degrees)
b1_map = ((meas_fa / 10) / requested_fa) * (np.pi / (GAMMA * 1e-3 * voltage_at_socket))
# Convert to [nT/V]
b1_map = b1_map * 1e9
# Save B1 map in [T/V] as NIfTI file
nii_b1 = nib.Nifti1Image(b1_map, nii.affine, nii.header)
nib.save(nii_b1, f"{subject}_TFLTB1map.nii.gz")
ref_voltage [V]: 387 (sub-CRMBM1_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-CRMBM1_acq-famp_TB1TFL)
ref_voltage [V]: 233 (sub-CRMBM2_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-CRMBM2_acq-famp_TB1TFL)
ref_voltage [V]: 380 (sub-CRMBM3_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-CRMBM3_acq-famp_TB1TFL)
ref_voltage [V]: 600 (sub-MGH1_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MGH1_acq-famp_TB1TFL)
ref_voltage [V]: 520 (sub-MGH2_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-MGH2_acq-famp_TB1TFL)
ref_voltage [V]: 635.7 (sub-MGH3_acq-famp_TB1TFL)
flip angle [degrees]: 88.94 (sub-MGH3_acq-famp_TB1TFL)
ref_voltage [V]: 649.6 (sub-MNI1_acq-famp_TB1TFL)
flip angle [degrees]: 86.87 (sub-MNI1_acq-famp_TB1TFL)
ref_voltage [V]: 446.4 (sub-MNI2_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MNI2_acq-famp_TB1TFL)
ref_voltage [V]: 595.5 (sub-MNI3_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-MNI3_acq-famp_TB1TFL)
ref_voltage [V]: 500 (sub-MPI1_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-MPI1_acq-famp_TB1TFL)
ref_voltage [V]: 510 (sub-MPI2_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MPI2_acq-famp_TB1TFL)
ref_voltage [V]: 510 (sub-MPI3_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-MPI3_acq-famp_TB1TFL)
ref_voltage [V]: 450 (sub-MSSM1_acq-famp_TB1TFL)
flip angle [degrees]: 60 (sub-MSSM1_acq-famp_TB1TFL)
ref_voltage [V]: 350 (sub-MSSM2_acq-famp_TB1TFL)
flip angle [degrees]: 80 (sub-MSSM2_acq-famp_TB1TFL)
ref_voltage [V]: 450 (sub-MSSM3_acq-famp_TB1TFL)
flip angle [degrees]: 60 (sub-MSSM3_acq-famp_TB1TFL)
ref_voltage [V]: 550 (sub-NTNU1_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-NTNU1_acq-famp_TB1TFL)
ref_voltage [V]: 450 (sub-NTNU2_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-NTNU2_acq-famp_TB1TFL)
ref_voltage [V]: 550 (sub-NTNU3_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-NTNU3_acq-famp_TB1TFL)
ref_voltage [V]: 388 (sub-UCL1_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-UCL1_acq-famp_TB1TFL)
ref_voltage [V]: 248 (sub-UCL2_acq-famp_TB1TFL)
flip angle [degrees]: 89.99 (sub-UCL2_acq-famp_TB1TFL)
ref_voltage [V]: 482 (sub-UCL3_acq-famp_TB1TFL)
flip angle [degrees]: 90 (sub-UCL3_acq-famp_TB1TFL)
Extract B1+ and SNR along the spinal cord#
# Extract B1+ and SNR along the spinal cord between levels C1 and T2 (included) and save data to CSV files
# TODO: remove code duplication
for subject in subjects:
os.chdir(os.path.join(path_data, subject, "fmap"))
# Extract TFL B1+ along the spinal cord
fname_result_b1plus = os.path.join(path_results, f"{subject}_TFLTB1map.csv")
!sct_extract_metric -i {subject}_TFLTB1map.nii.gz -f {subject}_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile {subject}_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o "{fname_result_b1plus}"
# Extract DREAM B1+ along the spinal cord
fname_result_b1plus = os.path.join(path_results, f"{subject}_DREAMTB1avgB1map.csv")
!sct_extract_metric -i {subject}_DREAMTB1avgB1map.nii.gz -f {subject}_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile {subject}_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o "{fname_result_b1plus}"
# Extract SNR along the spinal cord
fname_result_SNR = os.path.join(path_results, f"{subject}_SNRmap.csv")
!sct_extract_metric -i {subject}_acq-coilQaSagLarge_SNR_T0000.nii.gz -f {subject}_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile {subject}_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o "{fname_result_SNR}"
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM1_TFLTB1map.nii.gz -f sub-CRMBM1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM1_DREAMTB1avgB1map.nii.gz -f sub-CRMBM1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-CRMBM1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM2_TFLTB1map.nii.gz -f sub-CRMBM2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM2_DREAMTB1avgB1map.nii.gz -f sub-CRMBM2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-CRMBM2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM3_TFLTB1map.nii.gz -f sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM3_DREAMTB1avgB1map.nii.gz -f sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-CRMBM3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-CRMBM3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH1_TFLTB1map.nii.gz -f sub-MGH1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH1_DREAMTB1avgB1map.nii.gz -f sub-MGH1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MGH1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH2_TFLTB1map.nii.gz -f sub-MGH2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH2_DREAMTB1avgB1map.nii.gz -f sub-MGH2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MGH2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH3_TFLTB1map.nii.gz -f sub-MGH3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH3_DREAMTB1avgB1map.nii.gz -f sub-MGH3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MGH3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MGH3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI1_TFLTB1map.nii.gz -f sub-MNI1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI1_DREAMTB1avgB1map.nii.gz -f sub-MNI1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MNI1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI2_TFLTB1map.nii.gz -f sub-MNI2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI2_DREAMTB1avgB1map.nii.gz -f sub-MNI2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MNI2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI3_TFLTB1map.nii.gz -f sub-MNI3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI3_DREAMTB1avgB1map.nii.gz -f sub-MNI3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MNI3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MNI3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI1_TFLTB1map.nii.gz -f sub-MPI1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI1_DREAMTB1avgB1map.nii.gz -f sub-MPI1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MPI1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI2_TFLTB1map.nii.gz -f sub-MPI2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI2_DREAMTB1avgB1map.nii.gz -f sub-MPI2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MPI2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI3_TFLTB1map.nii.gz -f sub-MPI3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI3_DREAMTB1avgB1map.nii.gz -f sub-MPI3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MPI3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MPI3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM1_TFLTB1map.nii.gz -f sub-MSSM1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM1_DREAMTB1avgB1map.nii.gz -f sub-MSSM1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MSSM1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM2_TFLTB1map.nii.gz -f sub-MSSM2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM2_DREAMTB1avgB1map.nii.gz -f sub-MSSM2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MSSM2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM3_TFLTB1map.nii.gz -f sub-MSSM3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM3_DREAMTB1avgB1map.nii.gz -f sub-MSSM3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-MSSM3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-MSSM3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU1_TFLTB1map.nii.gz -f sub-NTNU1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU1_DREAMTB1avgB1map.nii.gz -f sub-NTNU1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-NTNU1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU2_TFLTB1map.nii.gz -f sub-NTNU2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU2_DREAMTB1avgB1map.nii.gz -f sub-NTNU2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-NTNU2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU3_TFLTB1map.nii.gz -f sub-NTNU3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU3_DREAMTB1avgB1map.nii.gz -f sub-NTNU3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-NTNU3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-NTNU3_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL1_TFLTB1map.nii.gz -f sub-UCL1_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL1_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL1_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL1_DREAMTB1avgB1map.nii.gz -f sub-UCL1_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL1_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL1_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL1_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_SNRmap.csv
--
Load metric image...
Estimation for label: sub-UCL1_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL1_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL2_TFLTB1map.nii.gz -f sub-UCL2_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL2_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL2_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL2_DREAMTB1avgB1map.nii.gz -f sub-UCL2_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL2_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL2_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL2_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_SNRmap.csv
--
Load metric image...
Estimation for label: sub-UCL2_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL2_SNRmap.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL3_TFLTB1map.nii.gz -f sub-UCL3_acq-anat_TB1TFL_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_TFLTB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL3_acq-anat_TB1TFL_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_TFLTB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL3_DREAMTB1avgB1map.nii.gz -f sub-UCL3_acq-famp_TB1DREAM_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_DREAMTB1avgB1map.csv
--
Load metric image...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Estimation for label: sub-UCL3_acq-famp_TB1DREAM_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_DREAMTB1avgB1map.csv
--
Spinal Cord Toolbox (6.5)
sct_extract_metric -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -f sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -method wa -vert 1:9 -vertfile sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -perlevel 1 -o /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_SNRmap.csv
--
Load metric image...
Estimation for label: sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg
Done! To view results, type:
xdg-open /home/runner/work/coil-qc-code/coil-qc-code/data-human/derivatives/results/sub-UCL3_SNRmap.csv
Generate plots along spinal cord#
Load and post-process extracted metrics#
# Reorder subjects so that identical coils are displayed side-by-side
subjects = ['sub-CRMBM1', 'sub-CRMBM2', 'sub-CRMBM3', 'sub-UCL1', 'sub-UCL2', 'sub-UCL3', 'sub-MNI1', 'sub-MNI2', 'sub-MNI3', 'sub-MGH1', 'sub-MGH2', 'sub-MGH3', 'sub-MPI1', 'sub-MPI2', 'sub-MPI3', 'sub-NTNU1', 'sub-NTNU2', 'sub-NTNU3', 'sub-MSSM1', 'sub-MSSM2', 'sub-MSSM3']
# Go back to root data folder
os.chdir(os.path.join(path_data))
def smooth_data(data, window_size=20):
""" Apply a simple moving average to smooth the data. """
return uniform_filter1d(data, size=window_size, mode='nearest')
# Fixed grid for x-axis
x_grid = np.linspace(0, 1, 100)
# z-slices corresponding to levels C3 to T2 on the PAM50 template. These will be used to scale the x-label of each subject.
original_vector = np.array([985, 939, 907, 870, 833, 800, 769, 735, 692, 646])
# Normalize the PAM50 z-slice numbers to the 1-0 range (to show inferior-superior instead of superior-inferior)
min_val = original_vector.min()
max_val = original_vector.max()
normalized_vector = 1 - ((original_vector - min_val) / (max_val - min_val))
# Use this normalized vector as x-ticks
custom_xticks = normalized_vector
# Vertebral level labels
vertebral_levels = ["C1", "C2", "C3", "C4", "C5", "C6", "C7", "T1", "T2"]
# Calculate midpoints for label positions
label_positions = normalized_vector[:-1] + np.diff(normalized_vector) / 2
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
# map types
map_types = ["TFLTB1", "DREAMTB1avgB1", "SNR"]
# Data storage for statistics
TFLTB1_data_stats = []
DREAMTB1avgB1_data_stats = []
SNR_TB1_data_stats = []
data_stats = [TFLTB1_data_stats, DREAMTB1avgB1_data_stats, SNR_TB1_data_stats]
# Data storage
TFLTB1_data = {}
DREAMTB1avgB1_data = {}
SNR_data = {}
data = [TFLTB1_data, DREAMTB1avgB1_data, SNR_data]
for map_type, data_stats_type, data_type in zip(map_types,data_stats,data):
i = 0
j = 0
for site in sites:
data_type[site]={}
while i < (j+3):
os.chdir(os.path.join(path_data, f"{subjects[i]}", "fmap"))
# Initialize list to collect data for this subject
subject_data = []
file_csv = os.path.join(path_results, f"{subjects[i]}_{map_type}map.csv")
df = pd.read_csv(file_csv)
wa_data = df['WA()']
# Compute stats on the non-resampled data (to avoid interpolation errors)
mean_data = np.mean(wa_data)
sd_data = np.std(wa_data)
# Normalize the x-axis to a 1-0 scale for each subject (to go from superior-inferior direction)
x_subject = np.linspace(1, 0, len(wa_data))
# Interpolate to the fixed grid
interp_func = interp1d(x_subject, wa_data, kind='linear', bounds_error=False, fill_value='extrapolate')
resampled_data = interp_func(x_grid)
# Apply smoothing
smoothed_data = smooth_data(resampled_data)
subject_data.append(smoothed_data)
for resampled_data in subject_data:
data_type[site][subjects[i]]=resampled_data
data_stats_type.append([site, subjects[i], mean_data, sd_data])
i += 1
j += 3
Normalize slice average SNR values by slice-average TFL B1+#
# Following https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.27695; here we normalize SNR maps by sin(FA), where FA is the actual FA map for the GRE SNR scan
# By normalizing we extrapolate to the SNR value that we would achive with FA = 90, ie, SNR_90 = SNR_meas/sin(FA_meas), which can be directly compated between RF coils
# Since we do not have the FA_meas (for the SNR GRE scan), we will compute it from the ratio of the measured and requested FA in the TFL B1+ scan
# FA_gre_meas = FA_gre_requested * (FA_TFL_meas/FA_TFL_requested)
# FA_TFL_meas/FA_TFL_requested was previously computed to obtain the TFL B1+ efficiency (stored in "data"), we will reconvert TFL B1+ efficiency to the FA_TFL_meas/FA_TFL_requested ratio
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
# SNR along the cord is stored in a dictionary (data) of dictionaries (?) that has an array associated with each element
# The order of the dictionary is {'CRMBM': {'sub-CRMBM1': array([...]), 'sub-CRMBM2': array([...]), etc.
# The 1st dictionary corresponds to the TFL B1+ data
# The 3rd/last dictionary corresponds to the SNR data
TFLB1_data = data[0]
SNR_data = data[2]
GAMMA = 2.675e8; # [rad / (s T)]
j = 0
i = 0
for site in sites:
while i < (j+3):
os.chdir(os.path.join(path_data, f"{subjects[i]}", "fmap"))
if subjects[i]=='sub-MSSM1':
ref_voltage=450
elif subjects[i]=='sub-MSSM2':
ref_voltage=350
elif subjects[i]=='sub-MSSM3':
ref_voltage=450
else:
# Fetch the reference voltage from the JSON sidecar
with open(f"{subjects[i]}_acq-famp_TB1TFL.json", "r") as f:
metadata = json.load(f)
ref_voltage = metadata.get("TxRefAmp", "N/A")
if (ref_voltage == "N/A"):
ref_token = "N/A"
for token in metadata.get("SeriesDescription", "N/A").split("_"):
if token.startswith("RefV"): ref_token = token
ref_voltage = float(ref_token[4:-1])
# Account for the power loss between the coil and the socket. That number was given by Siemens.
voltage_at_socket = ref_voltage * 10 ** -0.095
# Fetch the requested flip angle for the SNR(GRE) scan from the JSON sidecar
with open(f"{subjects[i]}_acq-coilQaSagLarge_SNR.json", "r") as f:
metadata = json.load(f)
requested_fa = metadata.get("FlipAngle", "N/A")
# compute the actual flip angle for the SNR(GRE) scan
actual_fa = requested_fa * (TFLB1_data[site][subjects[i]]/1e9) * ((GAMMA * 1e-3 * voltage_at_socket)/np.pi)
# normalize the SNR data by the actual flip angle
data[2][site][subjects[i]] = SNR_data[site][subjects[i]]/np.sin(np.deg2rad(actual_fa))
i += 1
j += 3
# replace SNR SC average and std in data_stats with SNR90 data
j = 0
i = 0
k = 0
for site in sites:
while i < (j+3):
data_stats[2][k][2] = np.mean(data[2][site][subjects[i]])
data_stats[2][k][3] = np.std(data[2][site][subjects[i]])
k += 1
i += 1
j += 3
Generate plots of B1+ and SNR along the cord#
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
site_colors = ['cornflowerblue', 'royalblue', 'firebrick', 'darkred', 'limegreen', 'green', 'mediumpurple']
subject_names = ["Subject 1", "Subject 2", "Subject 3", "average"]
sub_linestyles = ['dotted','dashed','dashdot']
sub_colors = ['dimgray','darkgray','silver']
# figure types
fig_types = ["TFL B1+ [nT/V]", "DREAM B1+ [nT/V]", "SNR_90"]
for data_type, data_stats_type, fig_type in zip(data, data_stats, fig_types):
avg_data = {'CRMBM': 0, 'UCL': 0, 'MNI': 0, 'MGH': 0, 'MPI': 0, 'NTNU': 0, 'MSSM': 0}
fig = plt.figure()
gs = fig.add_gridspec(1, len(sites), wspace=0)
axs = gs.subplots(sharex=True, sharey=True)
fig.set_size_inches(16, 6)
j = 0
i = 0
for k, site in enumerate(sites):
l = 0
while i < (j+3):
axs[k].plot(data_type[site][subjects[i]],color=sub_colors[l], linestyle=sub_linestyles[l])
avg_data[site] += data_type[site][subjects[i]]
l += 1
i += 1
j += 3
avg_data[site] = avg_data[site]/3
axs[k].plot(avg_data[site],color="black",linestyle='solid',linewidth=1)
axs[k].set_title(sites[k], color=site_colors[k])
axs[k].grid()
axs[0].legend(subject_names,loc="upper right")
for ax in axs.flat:
ax.set(xlabel='Vertebral Levels', ylabel=fig_type, xticks=100*label_positions, xticklabels=vertebral_levels)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45, ha='center')
if fig_type=="SNR_90":
ax.set_ylim(200, 1300)
else:
ax.set_ylim(0, 85)
# Hide x labels and tick labels for top plots and y ticks for right plots.
for ax in axs.flat:
ax.label_outer()
plt.show()
print('Subject-averaged', fig_type,'averaged along SC:',*zip(*{k: np.mean(v) for k, v in avg_data.items()}.items()),'\n')
print('Subject-averaged', fig_type,'std along SC:',*zip(*{k: np.std(v) for k, v in avg_data.items()}.items()),'\n')
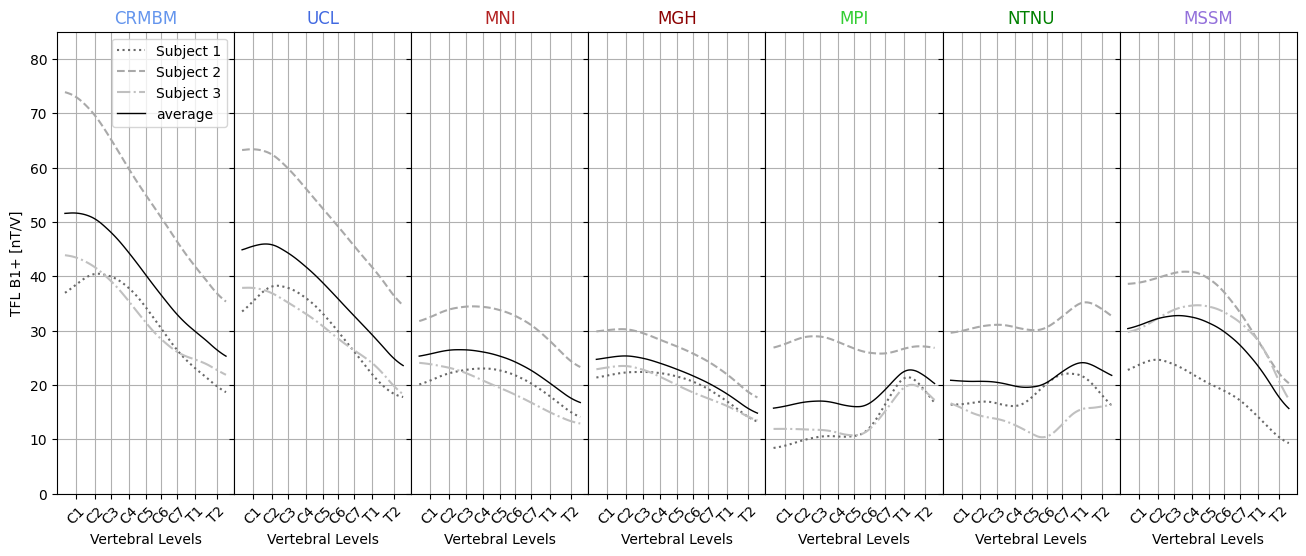
Subject-averaged TFL B1+ [nT/V] averaged along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(39.93356260254999), np.float64(37.39520489692691), np.float64(23.676592328930937), np.float64(21.882446479876236), np.float64(18.16152873545494), np.float64(21.302348080576603), np.float64(28.333119882266598))
Subject-averaged TFL B1+ [nT/V] std along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(9.009996302084597), np.float64(7.376676479079951), np.float64(3.0524725341725305), np.float64(3.2922307236237507), np.float64(2.3954373291064797), np.float64(1.4432340268800385), np.float64(5.050655094212115))
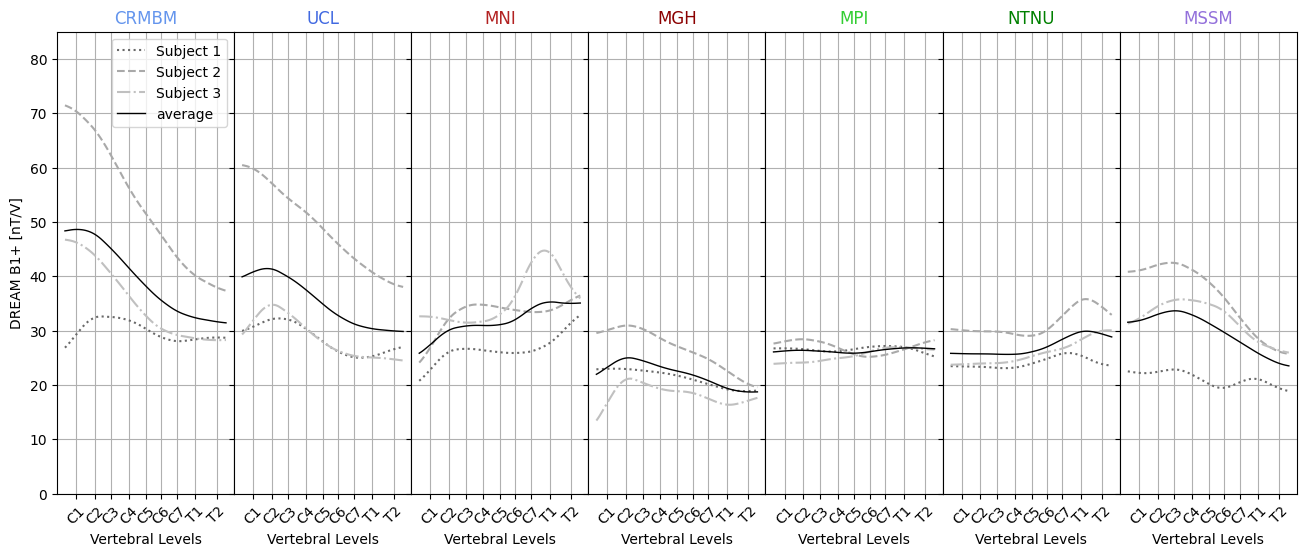
Subject-averaged DREAM B1+ [nT/V] averaged along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(39.36771115234862), np.float64(35.35388072501364), np.float64(31.837834436423876), np.float64(22.031627812816954), np.float64(26.33482955892221), np.float64(27.11647518125965), np.float64(29.83728997391403))
Subject-averaged DREAM B1+ [nT/V] std along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(6.416527213952659), np.float64(4.404232804428753), np.float64(2.603268756155278), np.float64(2.0888421628029454), np.float64(0.3126350638528332), np.float64(1.6456022597906872), np.float64(3.3436260992431053))
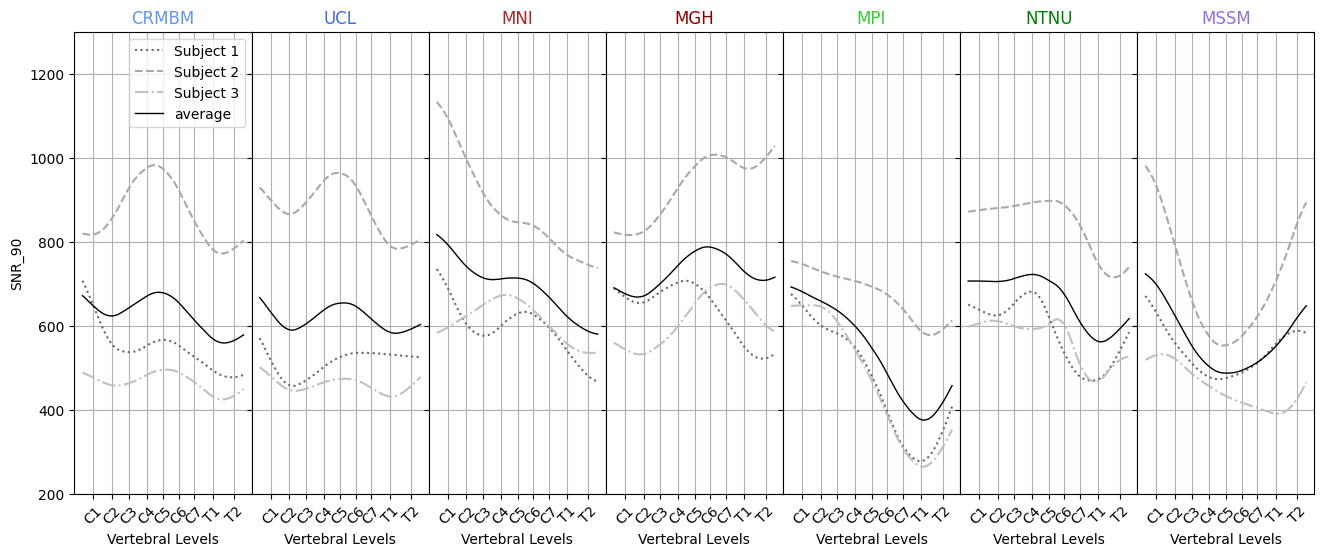
Subject-averaged SNR_90 averaged along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(627.0114196422146), np.float64(616.7775941543039), np.float64(693.6088225984932), np.float64(726.4827979145156), np.float64(535.2657270609408), np.float64(663.1814143787899), np.float64(566.8045632204831))
Subject-averaged SNR_90 std along SC: ('CRMBM', 'UCL', 'MNI', 'MGH', 'MPI', 'NTNU', 'MSSM') (np.float64(39.66589654462815), np.float64(25.40296498038003), np.float64(62.906550698018975), np.float64(39.75781841720328), np.float64(111.84441973828244), np.float64(58.8456618966059), np.float64(71.22951003335598))
# print some stats
# to do: automate this
print("CRMBM std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][0][2],data_stats[0][1][2],data_stats[0][2][2]]))
print("UCL std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][3][2],data_stats[0][4][2],data_stats[0][5][2]]))
print("MNI std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][6][2],data_stats[0][7][2],data_stats[0][8][2]]))
print("MGH std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][9][2],data_stats[0][10][2],data_stats[0][11][2]]))
print("MPI std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][12][2],data_stats[0][13][2],data_stats[0][14][2]]))
print("NTNU std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][15][2],data_stats[0][16][2],data_stats[0][17][2]]))
print("MSSM std across subjects of mean TFL B1+ along SC: ",np.std([data_stats[0][18][2],data_stats[0][19][2],data_stats[0][20][2]]))
print("\n")
print("CRMBM std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][0][2],data_stats[1][1][2],data_stats[1][2][2]]))
print("UCL std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][3][2],data_stats[1][4][2],data_stats[1][5][2]]))
print("MNI std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][6][2],data_stats[1][7][2],data_stats[1][8][2]]))
print("MGH std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][9][2],data_stats[1][10][2],data_stats[1][11][2]]))
print("MPI std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][12][2],data_stats[1][13][2],data_stats[1][14][2]]))
print("NTNU std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][15][2],data_stats[1][16][2],data_stats[1][17][2]]))
print("MSSM std across subjects of mean DREAM B1+ along SC: ",np.std([data_stats[1][18][2],data_stats[1][19][2],data_stats[1][20][2]]))
print("\n")
print("CRMBM std across subjects: ",np.mean([np.std([data_stats[0][0][2],data_stats[0][1][2],data_stats[0][2][2]]),np.std([data_stats[1][0][2],data_stats[1][1][2],data_stats[1][2][2]])]))
print("UCL std across subjects: ",np.mean([np.std([data_stats[0][3][2],data_stats[0][4][2],data_stats[0][5][2]]),np.std([data_stats[1][3][2],data_stats[1][4][2],data_stats[1][5][2]])]))
print("MNI std across subjects: ",np.mean([np.std([data_stats[0][6][2],data_stats[0][7][2],data_stats[0][8][2]]),np.std([data_stats[1][6][2],data_stats[1][7][2],data_stats[1][8][2]])]))
print("MGH std across subjects: ",np.mean([np.std([data_stats[0][9][2],data_stats[0][10][2],data_stats[0][11][2]]),np.std([data_stats[1][9][2],data_stats[1][10][2],data_stats[1][11][2]])]))
print("MPI std across subjects: ",np.mean([np.std([data_stats[0][12][2],data_stats[0][13][2],data_stats[0][14][2]]),np.std([data_stats[1][12][2],data_stats[1][13][2],data_stats[1][14][2]])]))
print("NTNU std across subjects: ",np.mean([np.std([data_stats[0][15][2],data_stats[0][16][2],data_stats[0][17][2]]),np.std([data_stats[1][15][2],data_stats[1][16][2],data_stats[1][17][2]])]))
print("MSSM std across subjects: ",np.mean([np.std([data_stats[0][18][2],data_stats[0][19][2],data_stats[0][20][2]]),np.std([data_stats[1][18][2],data_stats[1][19][2],data_stats[1][20][2]])]))
CRMBM std across subjects of mean TFL B1+ along SC: 10.867004066007558
UCL std across subjects of mean TFL B1+ along SC: 10.09553696735376
MNI std across subjects of mean TFL B1+ along SC: 5.396384432808582
MGH std across subjects of mean TFL B1+ along SC: 2.94241423550438
MPI std across subjects of mean TFL B1+ along SC: 6.417458801845382
NTNU std across subjects of mean TFL B1+ along SC: 7.49483466983962
MSSM std across subjects of mean TFL B1+ along SC: 6.551542107044434
CRMBM std across subjects of mean DREAM B1+ along SC: 9.892631531013066
UCL std across subjects of mean DREAM B1+ along SC: 9.58656441041675
MNI std across subjects of mean DREAM B1+ along SC: 3.7625276905355896
MGH std across subjects of mean DREAM B1+ along SC: 3.441573734373501
MPI std across subjects of mean DREAM B1+ along SC: 0.6670820484605456
NTNU std across subjects of mean DREAM B1+ along SC: 3.096226309213366
MSSM std across subjects of mean DREAM B1+ along SC: 6.242806779790072
CRMBM std across subjects: 10.379817798510313
UCL std across subjects: 9.841050688885256
MNI std across subjects: 4.579456061672086
MGH std across subjects: 3.1919939849389403
MPI std across subjects: 3.5422704251529638
NTNU std across subjects: 5.295530489526493
MSSM std across subjects: 6.397174443417253
Generate plots of B1+ and SNR per vertebral levels#
# figure types
site_colors = ['cornflowerblue', 'royalblue', 'firebrick', 'darkred', 'limegreen', 'green', 'mediumpurple']
fig_types = ["TFL B1+ CoV [nT/V] across C1-T2", "DREAM B1+ CoV [nT/V] across C1-T2", "SNR_90 Mean [arb] across C1-T2"]
subject_names = ["Subject 1", "Subject 2", "Subject 3"]
sub_colors = ['dimgray','darkgray','silver']
for data_stats_type, fig_type in zip(data_stats,fig_types):
series = [data_stats_type[i::len(subject_names)] for i in range(len(subject_names))]
hline_x = np.arange(len(sites))
hline_width = 0.25
stat_metric = np.zeros((len(subject_names),len(sites)))
fig, ax = plt.subplots()
i = 0
for subject_name, subject_series in zip(subject_names, series):
if fig_type == "SNR_90 Mean [arb] across C1-T2":
# Compute mean across levels
metric_indiv = [subject_series[j][2] for j in range(len(sites))]
else:
# Compute CoV across levels
metric_indiv = [subject_series[j][3]/subject_series[j][2] for j in range(len(sites))]
ax.scatter(sites, metric_indiv, label=subject_name, color=sub_colors[i])
for xtick, site_color in zip(ax.get_xticklabels(), site_colors):
xtick.set_color(site_color)
for j in range(len(sites)):
if fig_type == "SNR_90 Mean [arb] across C1-T2":
# Compute mean across levels and subjects
stat_metric[i][j] = subject_series[j][2]
else:
# Compute CoV across levels and subjects
stat_metric[i][j] = subject_series[j][3]/subject_series[j][2]
i+=1
plt.hlines(np.mean(stat_metric, axis=0),hline_x - hline_width/2, hline_x + hline_width/2, color="black", label="Across subj. mean")
ax.legend()
if fig_type=="SNR_90 Mean [arb] across C1-T2":
ax.set_ylim(200, 1000)
#ax.get_legend().remove()
else:
ax.set_ylim(0, 0.5)
ax.set_title(fig_type)
plt.grid()
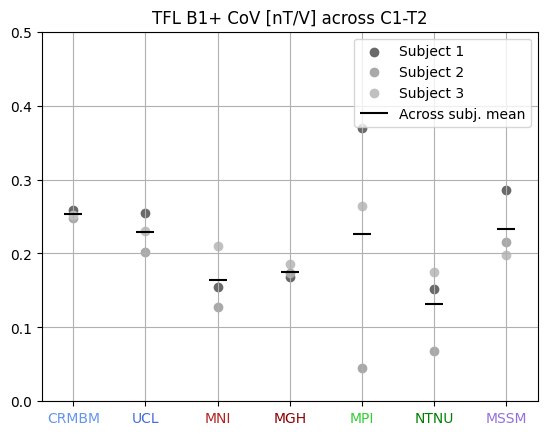
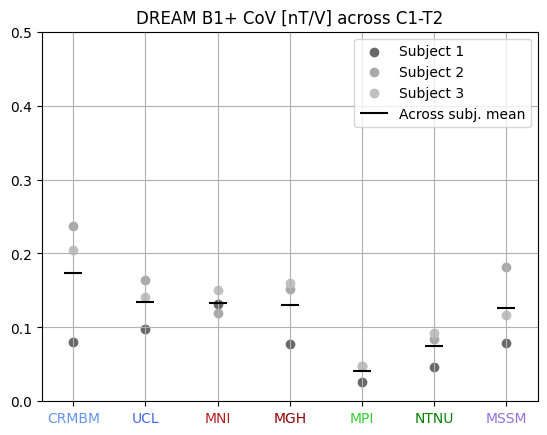
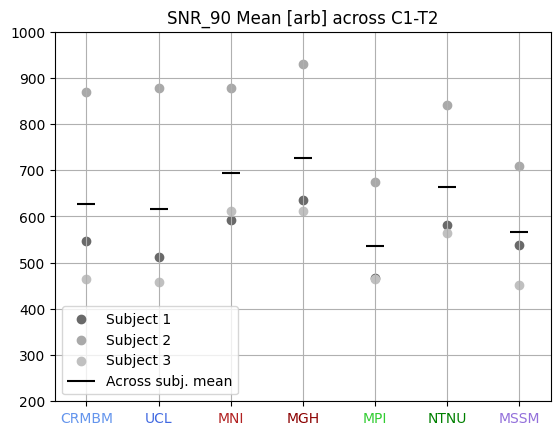
Generate maps#
Generate figures for B1+, SNR, and 1/g-factor maps obtained at each site (for one representative subject)
Co-register maps to reference site#
# Co-register subjects across sites for better visualisation
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
subject_id = '3' # Subject number to generate figures from
site_ref = 'CRMBM'
vert_labels = '3' # vertebral levels for alignment between sites
files_anat = ["acq-anat_TB1TFL", "acq-famp_TB1DREAM","acq-coilQaSagLarge_SNR_T0000"] # image that serves as a reference to get the segmentation
files_metric = ["TFLTB1map", "DREAMTB1avgB1map","acq-coilQaSagLarge_SNR_T0000"] # coilQA image to display on the figure
for site in sites:
print(f"👉 PROCESSING: {site}{subject_id}")
for file_anat, file_metric in zip(files_anat, files_metric):
os.chdir(os.path.join(path_data, "sub-"+site+subject_id, "fmap"))
# Extract vertebral labels
!sct_label_utils -i sub-{site}{subject_id}_{file_anat}_seg_labeled-UNIT1reg.nii.gz -vert-body {vert_labels} -o sub-{site}{subject_id}_{file_anat}_labels.nii.gz
# Co-register data to reference subject
if site != "CRMBM":
!sct_register_multimodal \
-i sub-{site}{subject_id}_{file_metric}.nii.gz \
-iseg sub-{site}{subject_id}_{file_anat}_seg.nii.gz \
-ilabel sub-{site}{subject_id}_{file_anat}_labels.nii.gz \
-d ../../sub-{site_ref}{subject_id}/fmap/sub-{site_ref}{subject_id}_{file_metric}.nii.gz \
-dseg ../../sub-{site_ref}{subject_id}/fmap/sub-{site_ref}{subject_id}_{file_anat}_seg.nii.gz \
-dlabel ../../sub-{site_ref}{subject_id}/fmap/sub-{site_ref}{subject_id}_{file_anat}_labels.nii.gz \
-param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 \
-x nn
👉 PROCESSING: CRMBM3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
👉 PROCESSING: UCL3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-UCL3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_TFLTB1map.nii.gz -iseg sub-UCL3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-UCL3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-UCL3_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-8.339592218399048, -9.86110806465149, 21.72777557373047]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 86
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[-10.52442002 -0.04140306 -8.54901552]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv/warp_src2dest.nii.gz warp_sub-UCL3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-UCL3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-UCL3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-UCL3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-16_register-wrapper_o2hjtrmv
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-UCL3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_DREAMTB1avgB1map.nii.gz -iseg sub-UCL3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-UCL3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-UCL3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-5.0303497314453125, -7.522422790527344, 20.21099853515625]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ -3.83318138 2.43617249 -11.59356689]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-UCL3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y/warp_src2dest.nii.gz warp_sub-UCL3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-UCL3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-UCL3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-UCL3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-20_register-wrapper_gnmnt31y
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-UCL3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-UCL3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-UCL3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-7.430351257324219, -9.022430419921875, 21.21099853515625]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 72
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ -6.33318138 -0.06384277 -10.09356689]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-UCL3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-UCL3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze/warp_src2dest.nii.gz warp_sub-UCL3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-UCL3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-UCL3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-23_register-wrapper_v5ed6fze
Finished! Elapsed time: 2s
👉 PROCESSING: MNI3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MNI3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_TFLTB1map.nii.gz -iseg sub-MNI3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-MNI3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MNI3_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.5, -60.16667032241821, 25.972225666046143]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 79
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[ 0.3151722 -50.34696531 -4.30456543]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds/warp_src2dest.nii.gz warp_sub-MNI3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-MNI3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-MNI3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-MNI3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-27_register-wrapper_frk3ssds
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MNI3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_DREAMTB1avgB1map.nii.gz -iseg sub-MNI3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-MNI3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MNI3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-0.0, -58.5, 27.5]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ 1.19716835 -48.54140472 -4.30456543]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MNI3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha/warp_src2dest.nii.gz warp_sub-MNI3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MNI3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MNI3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MNI3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-31_register-wrapper_jj5gioha
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MNI3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-MNI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-MNI3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-1.9073486328125e-06, -61.0, 27.0]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ 1.09716797 -52.04141235 -4.30456543]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MNI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MNI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en/warp_src2dest.nii.gz warp_sub-MNI3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MNI3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MNI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-33_register-wrapper_6kl528en
Finished! Elapsed time: 2s
👉 PROCESSING: MGH3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MGH3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_TFLTB1map.nii.gz -iseg sub-MGH3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-MGH3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MGH3_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-3.120776653289795, -73.15208745002747, 33.21037411689758]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[ -5.30560446 -63.33238244 2.93358302]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1/warp_src2dest.nii.gz warp_sub-MGH3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-MGH3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-MGH3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-MGH3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-37_register-wrapper_uo5xddb1
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MGH3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_DREAMTB1avgB1map.nii.gz -iseg sub-MGH3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-MGH3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MGH3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-5.2213897705078125, -72.7152099609375, 34.10382080078125]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ -4.02422142 -62.75661469 2.29925537]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MGH3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_/warp_src2dest.nii.gz warp_sub-MGH3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MGH3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MGH3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MGH3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-41_register-wrapper_b0u17mj_
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MGH3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-MGH3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-MGH3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz (255, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-6.600725173950195, -72.59494018678092, 33.02645874107911]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 49
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ -5.5035553 -63.63635254 1.72189331]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MGH3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MGH3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t/warp_src2dest.nii.gz warp_sub-MGH3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MGH3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MGH3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-44_register-wrapper_07n79j7t
Finished! Elapsed time: 2s
👉 PROCESSING: MPI3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MPI3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_TFLTB1map.nii.gz -iseg sub-MPI3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-MPI3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MPI3_TFLTB1map.nii.gz (90, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-6.205670356750488, -71.6954779624939, 26.370842456817627]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[ -8.39049816 -61.87577295 -3.90594864]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy/warp_src2dest.nii.gz warp_sub-MPI3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-MPI3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-MPI3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-MPI3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-48_register-wrapper_bnc_xgmy
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MPI3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_DREAMTB1avgB1map.nii.gz -iseg sub-MPI3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-MPI3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MPI3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-9.586259841918945, -71.69548034667969, 28.870834350585938]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ -8.38909149 -61.73688507 -2.93373108]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MPI3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_/warp_src2dest.nii.gz warp_sub-MPI3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MPI3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MPI3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MPI3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-51_register-wrapper_20mqzud_
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MPI3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-MPI3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-MPI3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz (255, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-7.086261749267578, -64.19546508789062, 27.370849609375]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ -5.98909187 -55.23687744 -3.93371582]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MPI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MPI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz/warp_src2dest.nii.gz warp_sub-MPI3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MPI3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MPI3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-54_register-wrapper_xp3krqhz
Finished! Elapsed time: 2s
👉 PROCESSING: NTNU3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-NTNU3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_TFLTB1map.nii.gz -iseg sub-NTNU3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-NTNU3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-NTNU3_TFLTB1map.nii.gz (88, 144, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[1.8448920249938965, -64.28926753997803, 38.56809759140015]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 80
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[ -0.33993578 -54.46956253 8.2913065 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd/warp_src2dest.nii.gz warp_sub-NTNU3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-NTNU3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-NTNU3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-NTNU3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-56-58_register-wrapper_5lt9j4jd
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-NTNU3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_DREAMTB1avgB1map.nii.gz -iseg sub-NTNU3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-NTNU3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-NTNU3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-1.5000553131103516, -62.90037536621094, 38.15143585205078]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 108
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ -0.30288696 -52.94178009 6.34687042]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-NTNU3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq/warp_src2dest.nii.gz warp_sub-NTNU3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-NTNU3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-NTNU3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-NTNU3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-57-02_register-wrapper_3cryiheq
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[-1.5000572204589844, -66.40036010742188, 39.15142822265625]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 3
Function evaluations: 72
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ -0.40288734 -57.44177246 7.84686279]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup/warp_src2dest.nii.gz warp_sub-NTNU3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-NTNU3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-NTNU3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-57-04_register-wrapper_suqz0uup
Finished! Elapsed time: 2s
👉 PROCESSING: MSSM3
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM3_acq-anat_TB1TFL_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MSSM3_acq-anat_TB1TFL_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_TFLTB1map.nii.gz -iseg sub-MSSM3_acq-anat_TB1TFL_seg.nii.gz -ilabel sub-MSSM3_acq-anat_TB1TFL_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MSSM3_TFLTB1map.nii.gz (88, 128, 56)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_TFLTB1map.nii.gz (88, 144, 56)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[5.0, -25.0, 35.0]]
Labels dest: [[2.1848278045654297, -9.81970500946045, 30.276791095733643]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[ 2.1848278 -9.81970501 30.2767911 ]
Translation:
[[ 2.8151722 -15.18029499 4.7232089 ]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9/warp_src2dest.nii.gz warp_sub-MSSM3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: warp_sub-MSSM3_TFLTB1map2sub-CRMBM3_TFLTB1map.nii.gz
File created: sub-CRMBM3_TFLTB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9/warp_dest2src.nii.gz warp_sub-CRMBM3_TFLTB1map2sub-MSSM3_TFLTB1map.nii.gz
File created: warp_sub-CRMBM3_TFLTB1map2sub-MSSM3_TFLTB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-57-08_register-wrapper_39u03ld9
Finished! Elapsed time: 2s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM3_acq-famp_TB1DREAM_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MSSM3_acq-famp_TB1DREAM_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_DREAMTB1avgB1map.nii.gz -iseg sub-MSSM3_acq-famp_TB1DREAM_seg.nii.gz -ilabel sub-MSSM3_acq-famp_TB1DREAM_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Source .............. sub-MSSM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_DREAMTB1avgB1map.nii.gz (80, 80, 11)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Creating temporary folder (/tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j)
Copying input data to tmp folder and convert to nii...
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.5, -25.5, 37.5]]
Labels dest: [[-1.1971683502197266, -9.958595275878906, 31.8045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.19716835 -9.95859528 31.80456543]
Translation:
[[ 3.69716835 -15.54140472 5.69543457]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File created: sub-MSSM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j/warp_src2dest.nii.gz warp_sub-MSSM3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-MSSM3_DREAMTB1avgB1map2sub-CRMBM3_DREAMTB1avgB1map.nii.gz
File created: sub-CRMBM3_DREAMTB1avgB1map_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j/warp_dest2src.nii.gz warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MSSM3_DREAMTB1avgB1map.nii.gz
File created: warp_sub-CRMBM3_DREAMTB1avgB1map2sub-MSSM3_DREAMTB1avgB1map.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-57-12_register-wrapper_itylfx0j
Finished! Elapsed time: 0s
--
Spinal Cord Toolbox (6.5)
sct_label_utils -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg_labeled-UNIT1reg.nii.gz -vert-body 3 -o sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz
--
Generating output files...
--
Spinal Cord Toolbox (6.5)
sct_register_multimodal -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -iseg sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -ilabel sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -d ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -dseg ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz -dlabel ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_labels.nii.gz -param step=0,type=label,dof=Tx_Ty_Tz:step=1,type=seg,iter=0 -x nn
--
Input parameters:
Source .............. sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (205, 256, 13)
Destination ......... ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz (256, 256, 13)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... label
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz
smoothWarpXY ... 2
rot_method ..... pca
Parameter 'algo=syn' has no effect for 'type=label' registration.
Labels src: [[2.3999977111816406, -26.499999955389995, 36.99999996831152]]
Labels dest: [[-1.0971698760986328, -8.958587646484375, 31.3045654296875]]
Degrees of freedom (dof): Tx_Ty_Tz
Optimization terminated successfully.
Current function value: 0.000000
Iterations: 2
Function evaluations: 50
Matrix:
[[ 1. 0. 0.]
[ 0. 1. 0.]
[-0. 0. 1.]]
Center:
[-1.09716988 -8.95858765 31.30456543]
Translation:
[[ 3.49716759 -17.54141231 5.69543454]]
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
Registration parameters:
type ........... seg
algo ........... syn
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/runner/sct_6.5/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MeanSquares[dest_seg_RPI.nii,src_seg_reg.nii,1,4]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step1,src_seg_reg_regStep1.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a
Concatenate transformations...
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_1.nii.gz warp_forward_0.txt # in /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a
/home/runner/sct_6.5/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_0.txt warp_inverse_1.nii.gz # in /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a
Apply transfo source --> dest...
Apply transfo dest --> source...
Generate output files...
File sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz already exists. Deleting it..
File created: sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a/warp_src2dest.nii.gz warp_sub-MSSM3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-MSSM3_acq-coilQaSagLarge_SNR_T00002sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz
mv /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a/warp_dest2src.nii.gz warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
File created: warp_sub-CRMBM3_acq-coilQaSagLarge_SNR_T00002sub-MSSM3_acq-coilQaSagLarge_SNR_T0000.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2025-03-20_02-57-15_register-wrapper_1n1gup8a
Finished! Elapsed time: 2s
Align spinal cord with the medial plane#
For visualization purpose, such that the sagittal views show the spinal cord throughout the whole superior-inferior axis.
# Flatten the spinal cord (ie, being it into the sagittal midline)
for site in sites:
print(f"👉 PROCESSING: {site}{subject_id}")
for file_anat, file_metric in zip(files_anat, files_metric):
os.chdir(os.path.join(path_data, "sub-"+site+subject_id, "fmap"))
if site == site_ref:
suffix = ""
else:
suffix = "_reg"
# Flatten the spinal cord in the sagittal plane
!sct_flatten_sagittal -i sub-{site}{subject_id}_{file_metric}{suffix}.nii.gz -s ../../sub-{site_ref}{subject_id}/fmap/sub-{site_ref}{subject_id}_{file_anat}_seg.nii.gz
👉 PROCESSING: CRMBM3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-CRMBM3_TFLTB1map.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-CRMBM3_DREAMTB1avgB1map.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'int16', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: UCL3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-UCL3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-UCL3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-UCL3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: MNI3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MNI3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MNI3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MNI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: MGH3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MGH3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MGH3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MGH3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: MPI3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MPI3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MPI3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MPI3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: NTNU3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-NTNU3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-NTNU3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-NTNU3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
👉 PROCESSING: MSSM3
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MSSM3_TFLTB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-anat_TB1TFL_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MSSM3_DREAMTB1avgB1map_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-famp_TB1DREAM_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'float64'. Header metadata will be overwritten to use 'float64'.
--
Spinal Cord Toolbox (6.5)
sct_flatten_sagittal -i sub-MSSM3_acq-coilQaSagLarge_SNR_T0000_reg.nii.gz -s ../../sub-CRMBM3/fmap/sub-CRMBM3_acq-coilQaSagLarge_SNR_T0000_seg.nii.gz
--
Image header specifies datatype 'float32', but array is of type 'int16'. Header metadata will be overwritten to use 'int16'.
Generate B1+ and SNR maps#
site_colors = ['cornflowerblue', 'royalblue', 'firebrick', 'darkred', 'limegreen', 'green', 'mediumpurple']
# legend types
legend_types = ["[nT/V]", "[nT/V]", "[arb]", "[arb]", "[arb]"]
# Define the slicing indices for each combination of file_metric and ind_subject
# The indices are defined as (x(min, max), y(min, max))
slicing_indices = {
"TFLTB1map": {
'1': (slice(31, 82), slice(43, 93)),
'2': (slice(28, 71), slice(37, 94)),
'3': (slice(25, 77), slice(35, 100))
},
"DREAMTB1avgB1map": {
'1': (slice(29, 60), slice(13, 66)),
'2': (slice(20, 65), slice(6, 56)),
'3': (slice(25, 72), slice(0, 60))
},
"acq-coilQaSagLarge_SNR_T0000": {
'1': (slice(224, 302), slice(213, 292)),
'2': (slice(224, 302), slice(197, 289)),
'3': (slice(100, 178), slice(55, 160))
}
}
# Define the dynmax values for each file_metric
dynmax_values = {
"TFLTB1map": 60,
"DREAMTB1avgB1map": 60,
"acq-coilQaSagLarge_SNR_T0000": 200
}
for file_metric, legend_type in zip(files_metric, legend_types):
# Create a figure with multiple subplots
fig, axes = plt.subplots(2, 4)#, figsize=(10, 8))
font_size = 12
axes=axes.flatten()
for i,site in enumerate(sites):
# Load data
os.chdir(os.path.join(path_data, f"sub-{site}{subject_id}", "fmap"))
if site == site_ref:
suffix = ""
else:
suffix = "_reg"
map = nib.load(f"sub-{site}{subject_id}_{file_metric}{suffix}_flatten.nii.gz")
# map = nib.load(f"sub-{site}{ind_subject}_{file_metric}{suffix}.nii.gz")
slices = slicing_indices[file_metric][subject_id]
data = map.get_fdata()[slices[0], slices[1], round(map.get_fdata().shape[2] / 2)]
# Figure configuration
axes[-1].axis('off')
# Defining dynamic range
axes[-1].axis('off')
dynmin = 0
dynmax = dynmax_values[file_metric]
splot=axes[i]
im = splot.imshow((data.T), cmap='viridis', origin='lower',vmin=dynmin,vmax=dynmax)
splot.set_title(site, size=font_size,color=site_colors[i])
splot.axis('off')
plt.tight_layout()
# Colorbar
# Assume that the colorbar should start at the bottom of the lower row of subplots and
# extend to the top of the upper row of subplots
cbar_bottom = 0.25 # This might need adjustment
cbar_height = 0.5 # This represents the total height of both rows of subplots
cbar_dist = 1.01
cbar_ax = fig.add_axes([cbar_dist, cbar_bottom, 0.03, cbar_height])
cbar = plt.colorbar(im, cax=cbar_ax)
cbar_ax.set_title(legend_type, size=12)
plt.show()
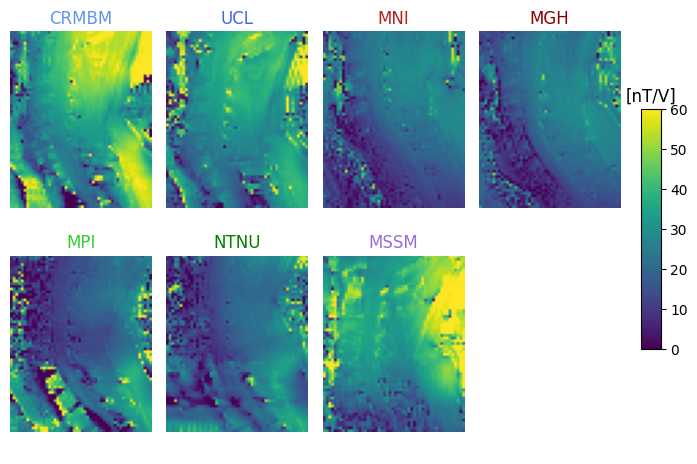
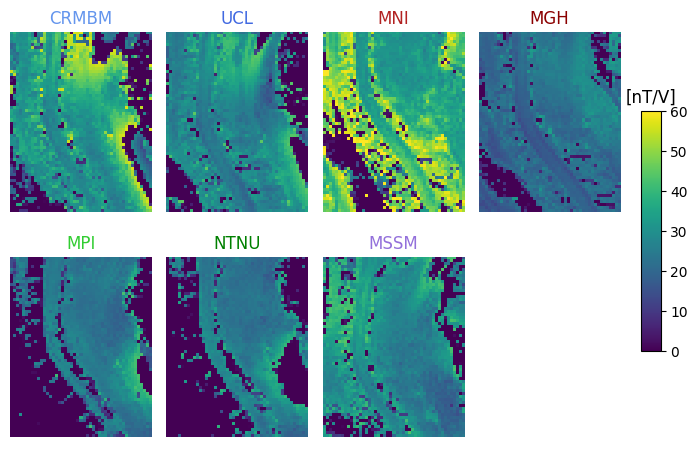

Generate g-factor maps#
# map types
map_types = ["acq-coilQaSagSmall_GFactor", "acq-coilQaTra_GFactor", "T2starw"]
sites = ["CRMBM", "MNI", "MPI", "MSSM", "UCL", "MGH", "NTNU"]
site_colors = ['cornflowerblue', 'firebrick', 'limegreen', 'mediumpurple', 'royalblue', 'darkred', 'green']
# legend types
legend_types = ["1/g", "1/g", "[arb]"]
# Select individual subject to show
ind_subject = '2' # Here we select subject 2 because it has the most complete data
mean_gfac = {}
max_gfac = {}
for map_type, legend_type in zip(map_types,legend_types):
# Create a figure with multiple subplots
fig, axes = plt.subplots(2, 4, figsize=(10, 8))
font_size = 12
axes=axes.flatten()
for i,site in enumerate(sites):
# Load data
if map_type=="T2starw":
os.chdir(os.path.join(path_data, f"sub-{site}{ind_subject}", "anat"))
map=nib.load(f"sub-{site}{ind_subject}_{map_type}.nii.gz")
data=map.get_fdata()[:,:,round(map.get_fdata().shape[2]/2)]
else:
os.chdir(os.path.join(path_data, f"sub-{site}{ind_subject}", "fmap"))
map=nib.load(f"sub-{site}{ind_subject}_{map_type}.nii.gz")
# The 4th dimension inlcudes 12 acceleration maps:
#['R 2','R 3', 'R 4', 'R 6', 'R 8', 'R 2 x 2', 'R 2 x 3', 'R 3 x 2', 'R 3 x 3', 'R 3 x 4', 'R 4 x 3', 'R 4 x 4']
if map_type=="acq-coilQaSagSmall_GFactor":
# show the R = 2 x 2 map
data=(map.get_fdata()[64:191,64:191,round(map.get_fdata().shape[2]/2),0])/1000
gfac_data=(map.get_fdata()[round(map.get_fdata().shape[0]/2)-10:round(map.get_fdata().shape[0]/2)+10,round(map.get_fdata().shape[1]/2)-10:round(map.get_fdata().shape[1]/2)+10,round(map.get_fdata().shape[2]/2),5])
else:
# show the R = 2 map
data=(map.get_fdata()[192:574,192:574,round(map.get_fdata().shape[2]/2),0])/1000
gfac_data=(map.get_fdata()[round(map.get_fdata().shape[0]/2)-30:round(map.get_fdata().shape[0]/2)+30,round(map.get_fdata().shape[1]/2)-30:round(map.get_fdata().shape[1]/2)+30,round(map.get_fdata().shape[2]/2),0])
mean_gfac[site]=np.nanmean(gfac_data)/1000
max_gfac[site]=np.max(gfac_data)/1000
# Plot
splot=axes[i]
dynmin = 0
if map_type=="T2starw":
dynmax = 3000
axes[-1].axis('off')
else:
dynmax = 1
axes[-1].axis('off')
splot.text(0, 3, r'mean 1/g='+str(round(mean_gfac[site],4)), size=10)
if map_type=="acq-coilQaSagSmall_GFactor":
x = [data.shape[0]/2-10, data.shape[1]/2-10]
y = [data.shape[0]/2-10, data.shape[1]/2+10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2-10, data.shape[1]/2+10]
y = [data.shape[0]/2+10, data.shape[1]/2+10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+10, data.shape[1]/2+10]
y = [data.shape[0]/2+10, data.shape[1]/2-10]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+10, data.shape[1]/2-10]
y = [data.shape[0]/2-10, data.shape[1]/2-10]
splot.plot(x, y, color="black", linewidth=2)
else:
x = [data.shape[0]/2-30, data.shape[1]/2-30]
y = [data.shape[0]/2-30, data.shape[1]/2+30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2-30, data.shape[1]/2+30]
y = [data.shape[0]/2+30, data.shape[1]/2+30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+30, data.shape[1]/2+30]
y = [data.shape[0]/2+30, data.shape[1]/2-30]
splot.plot(x, y, color="black", linewidth=2)
x = [data.shape[0]/2+30, data.shape[1]/2-30]
y = [data.shape[0]/2-30, data.shape[1]/2-30]
splot.plot(x, y, color="black", linewidth=2)
im = splot.imshow((data.T), cmap='viridis', origin='lower',vmin=dynmin,vmax=dynmax)
splot.set_title(site, size=font_size,color=site_colors[i])
splot.axis('off')
plt.tight_layout()
# Colorbar
# Assume that the colorbar should start at the bottom of the lower row of subplots and
# extend to the top of the upper row of subplots
cbar_bottom = 0.25 # This might need adjustment
cbar_height = 0.5 # This represents the total height of both rows of subplots
cbar_dist = 1.01
cbar_ax = fig.add_axes([cbar_dist, cbar_bottom, 0.03, cbar_height])
cbar = plt.colorbar(im, cax=cbar_ax)
cbar_ax.set_title(legend_type, size=12)
plt.show()
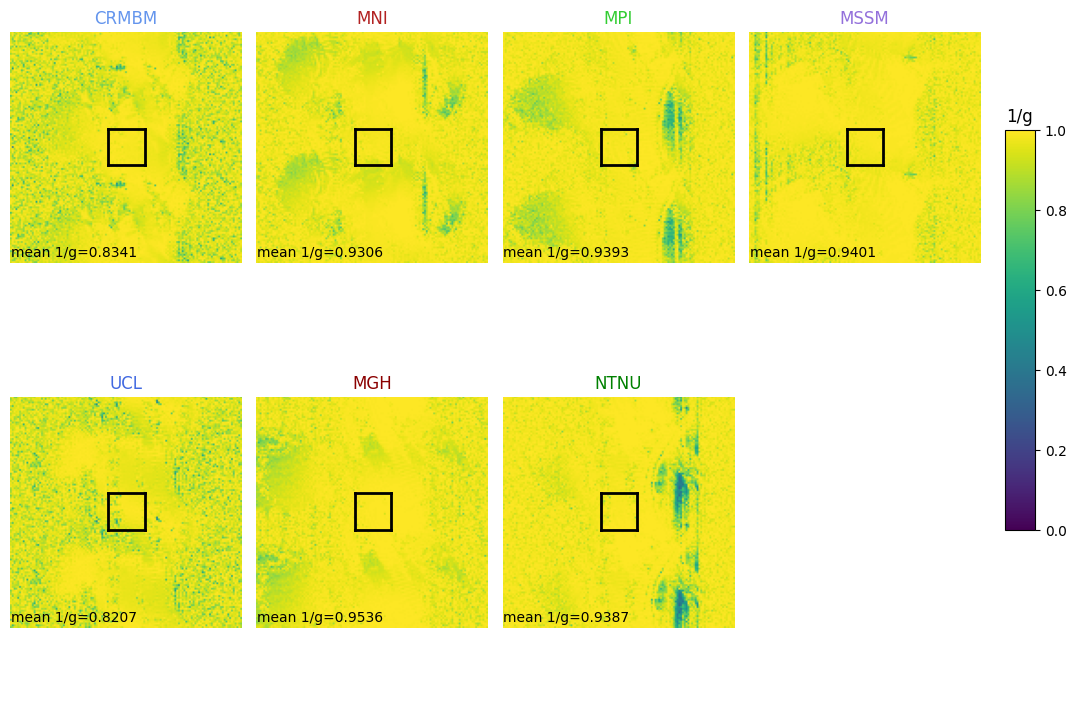
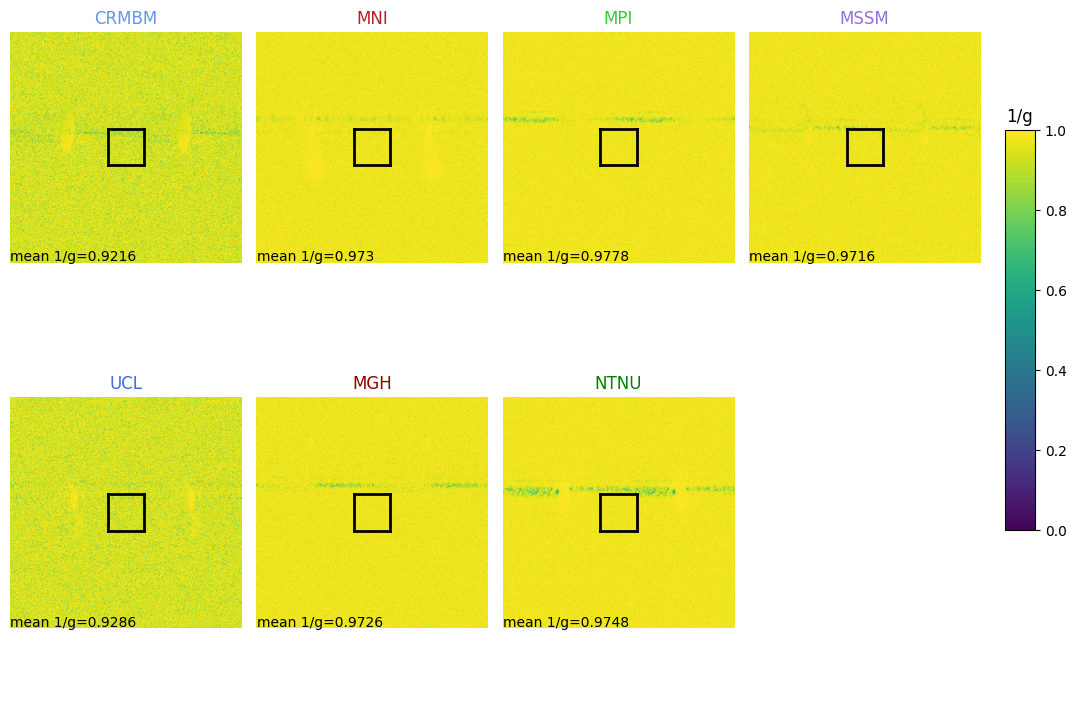
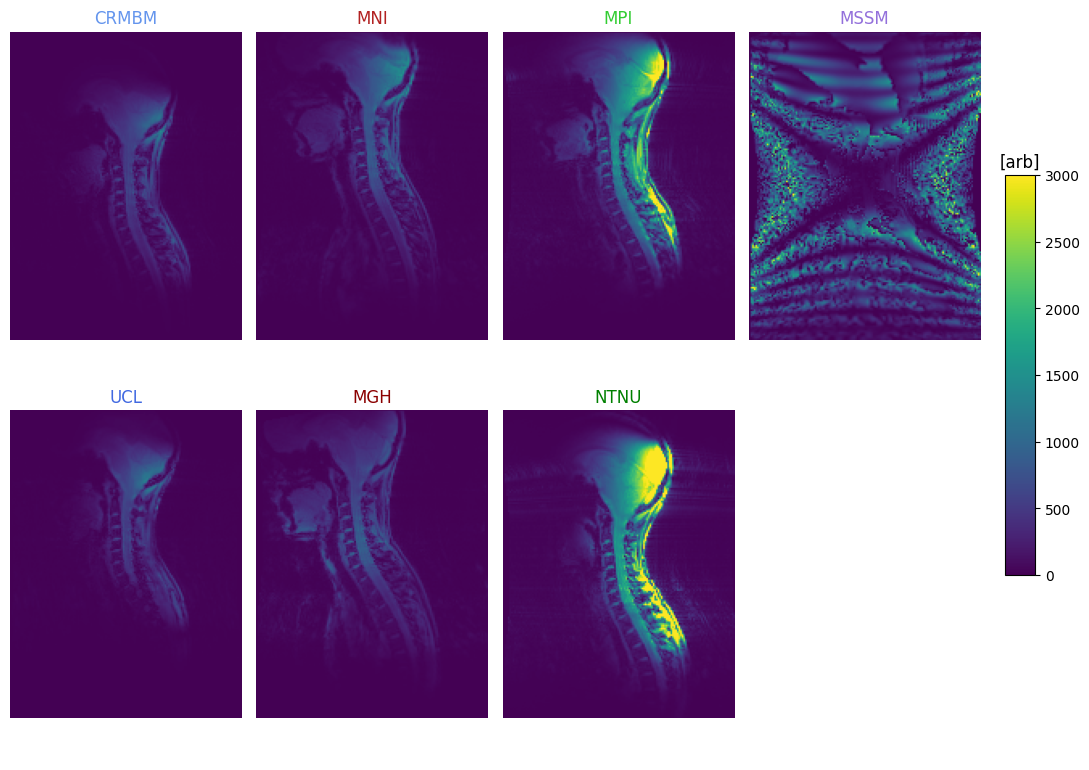
Generate tiled figure with individual channel on GRE scans#
sites = ["CRMBM", "UCL", "MNI", "MGH", "MPI", "NTNU", "MSSM"]
# Select subject to show
subject = '1' # and here we select subject 1 because it has the most complete data
for i,site in enumerate(sites):
gre_files=sorted(glob.glob(os.path.join(path_data, f"sub-{site}{subject}", "anat", '*uncombined*.nii.gz')))
#Tiled figure in a five-row layout
rows=int(np.ceil(len(gre_files)/4))
cols=int(np.ceil(len(gre_files)/rows))
fig=plt.figure(figsize=(15, 20))
ax = fig.subplots(rows,cols,squeeze=True)
for row in range(rows):
for col in range(cols):
i = row*cols+col
if i < len(gre_files):
#read in files
data_to_plot=(nib.load(gre_files[i])).get_fdata() #load in nifti object, get only image data
data_to_plot=np.rot90(data_to_plot[:,:,int(np.floor(data_to_plot.shape[2]/2))]) #central slice
ax[row,col].imshow(data_to_plot,cmap=plt.cm.gray,clim=[0, 300])
ax[row,col].text(0.5, 0.05, 'Rx channel : ' + str(i+1),horizontalalignment='center', transform=ax[row,col].transAxes,color='white',fontsize=17)
ax[row,col].axis('off')
plt.axis('off')
plt.subplots_adjust(hspace=0,wspace=0)
fig.suptitle(site, fontsize=20, y=0.9)
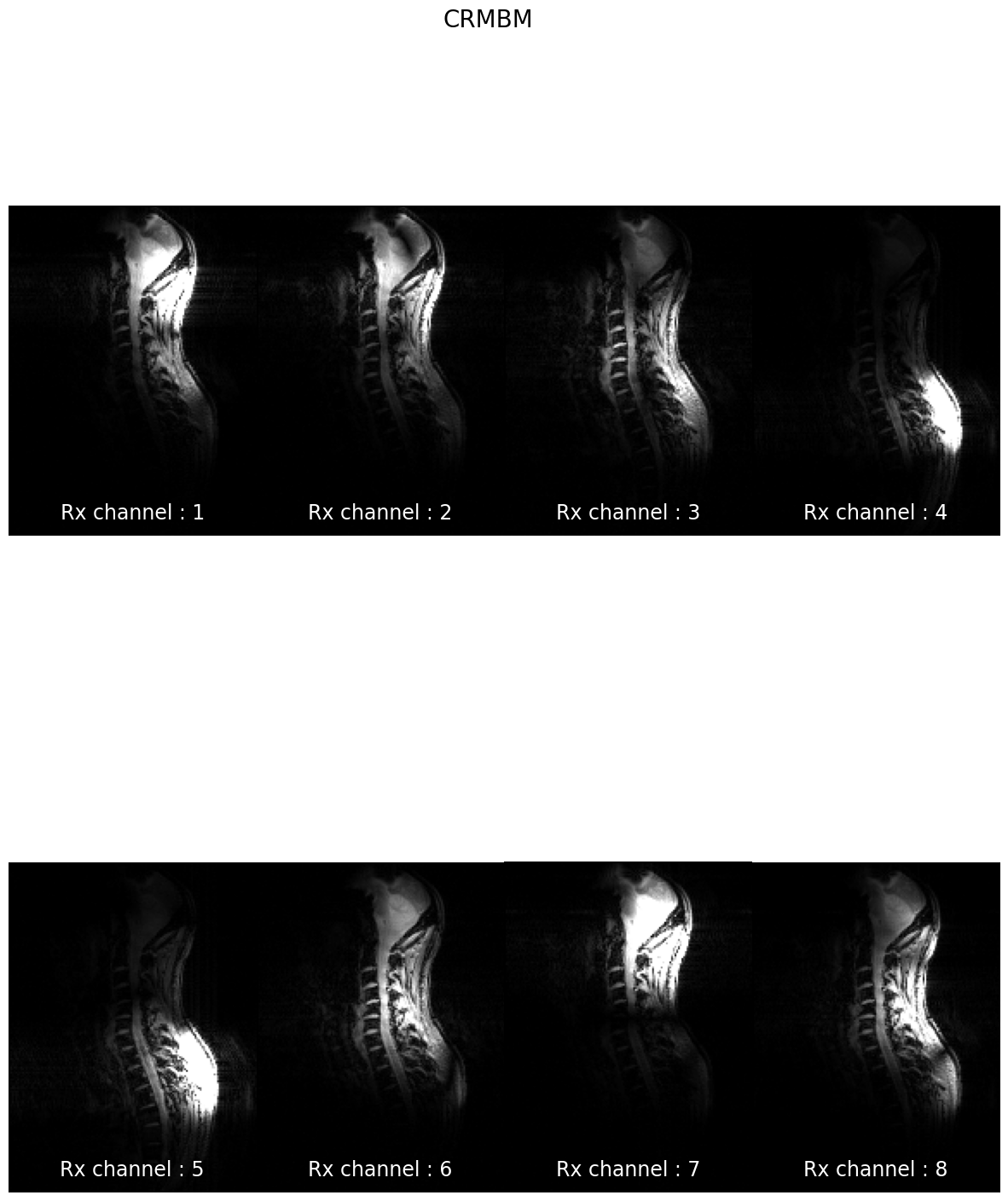
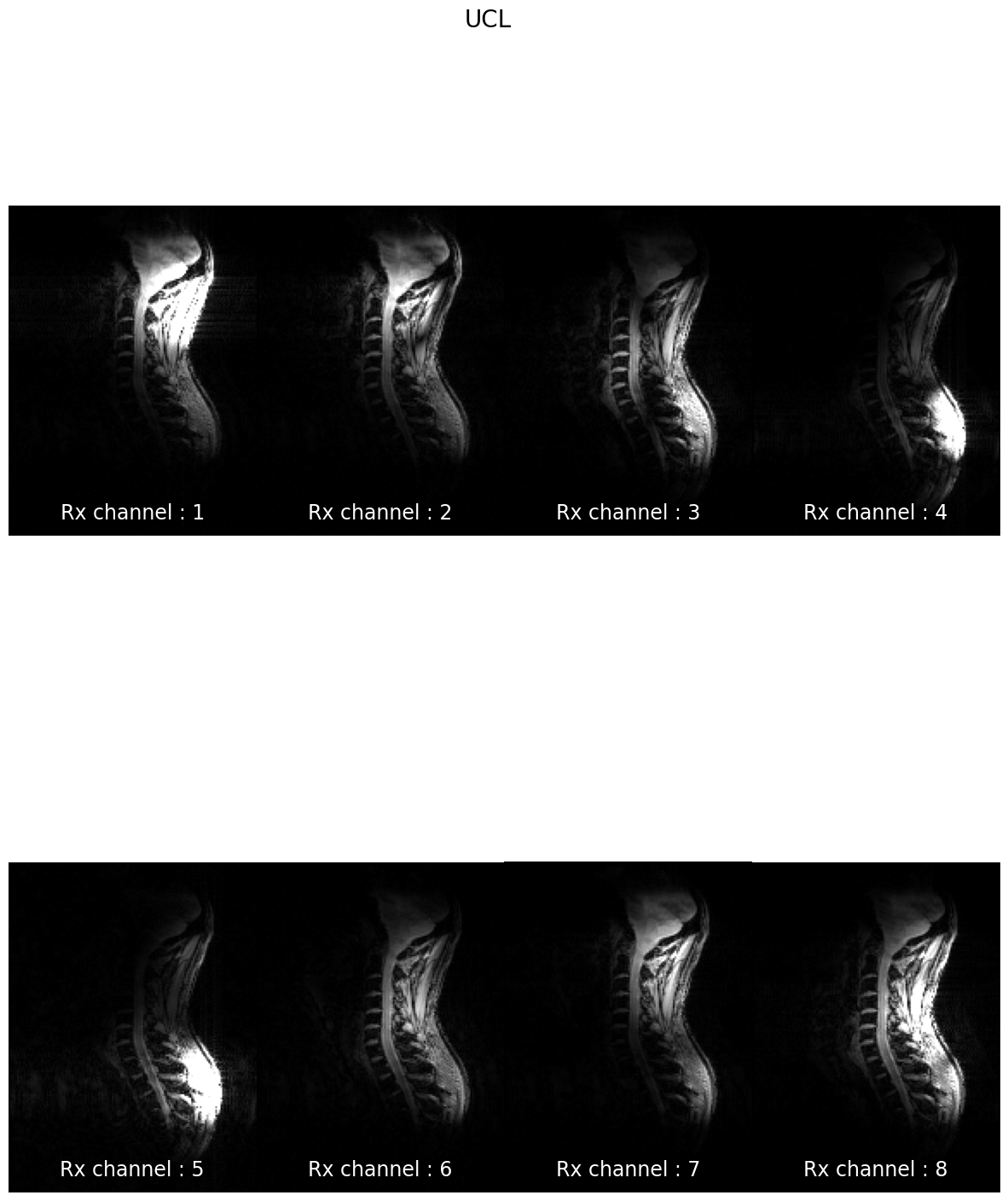
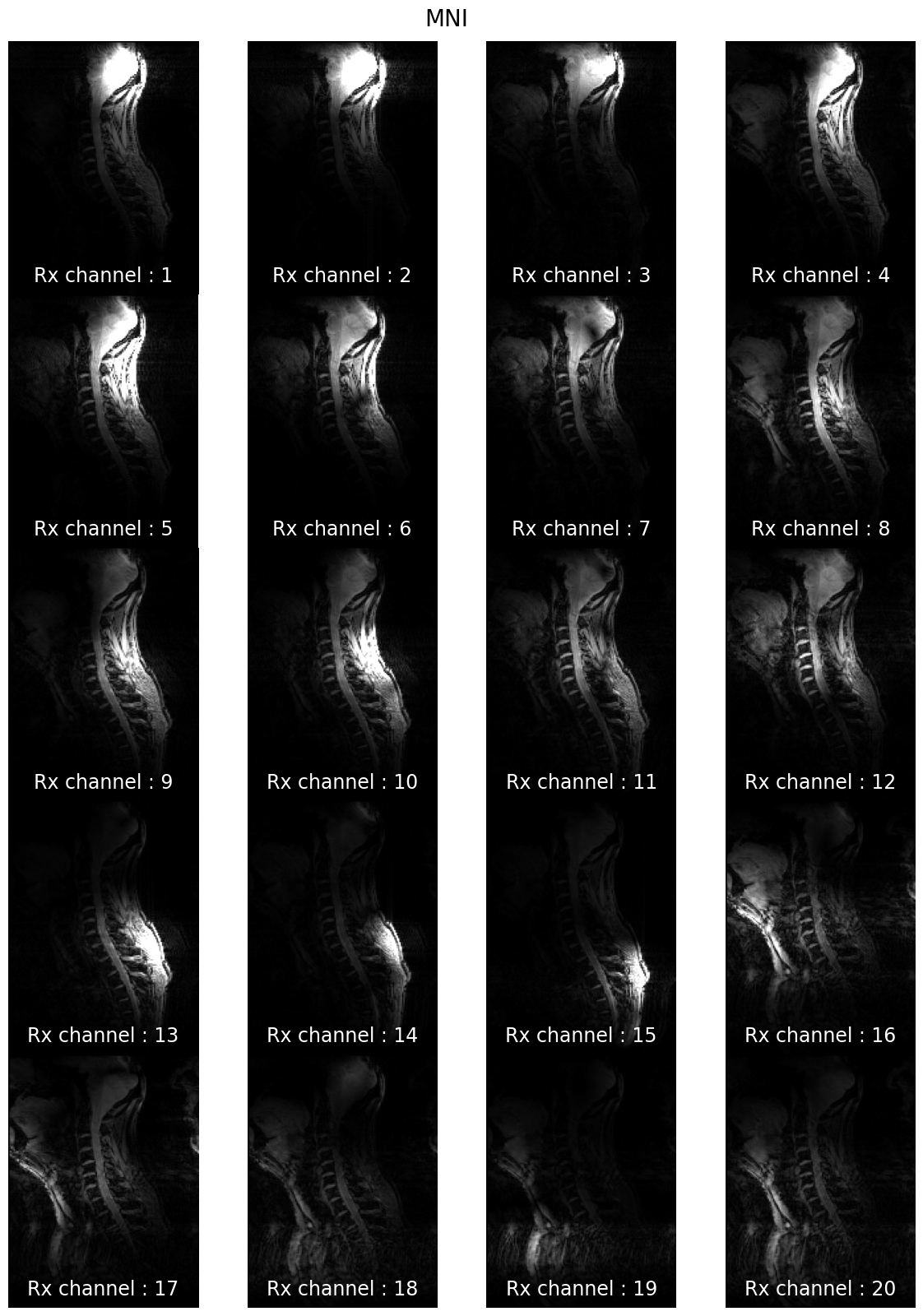
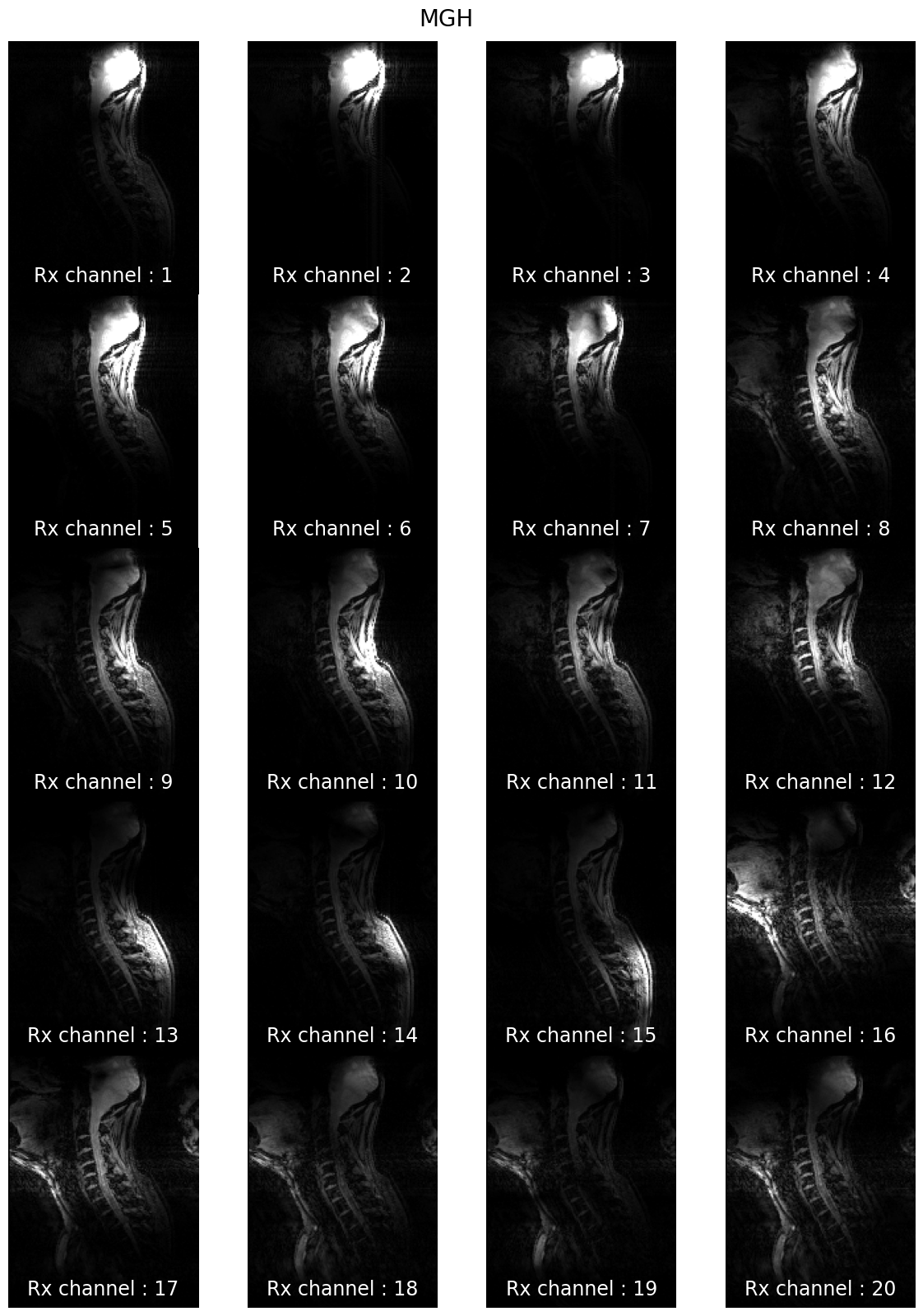
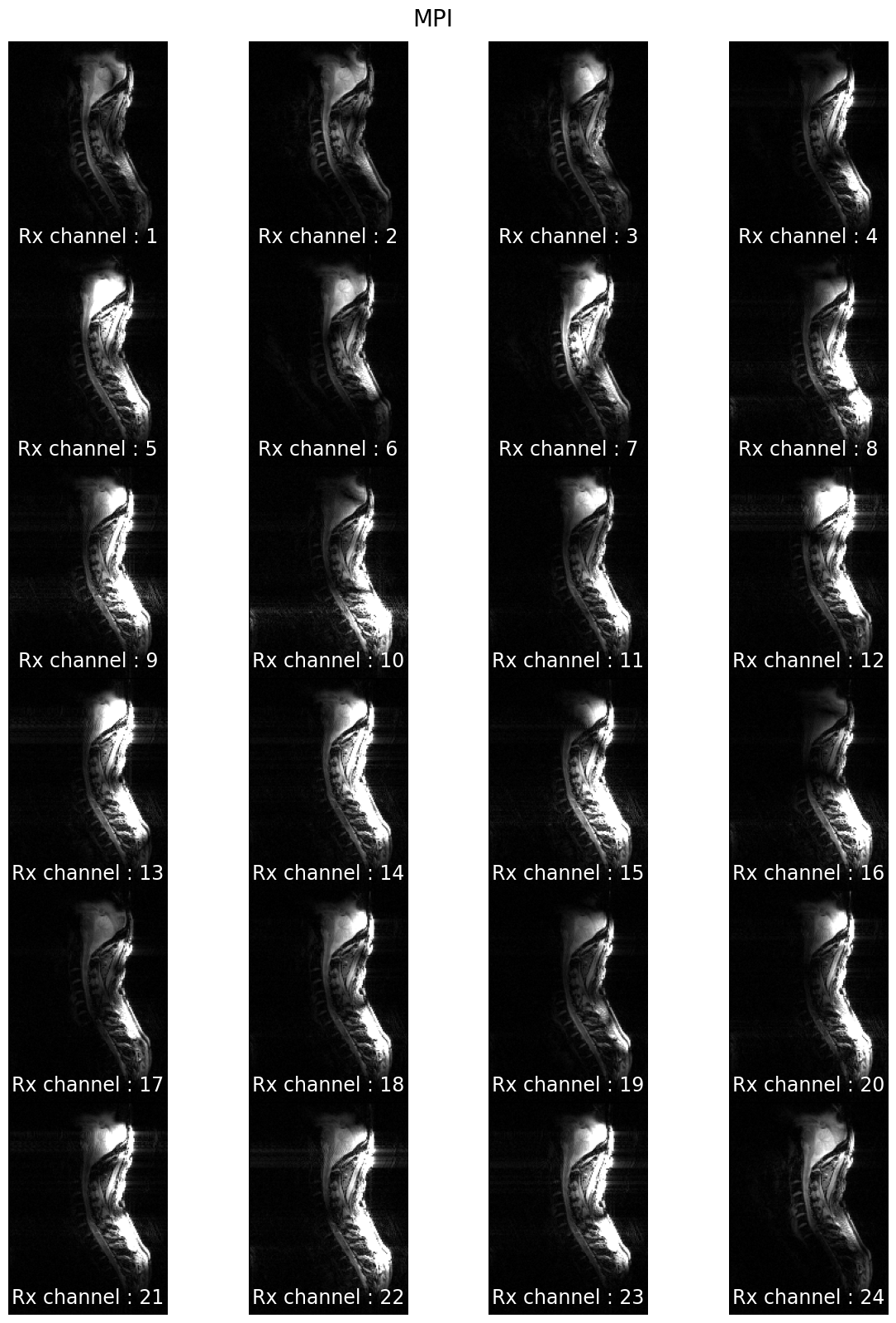
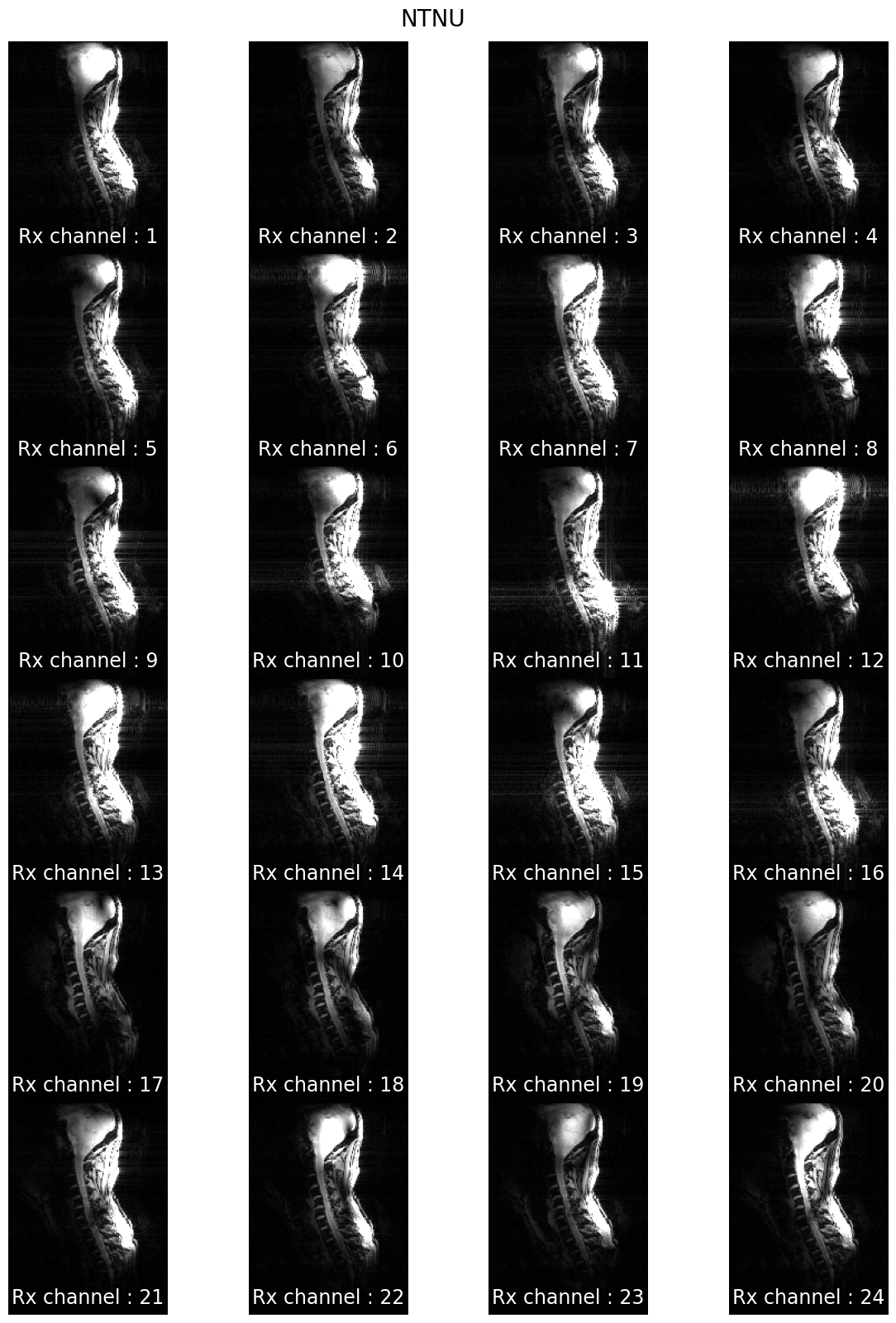
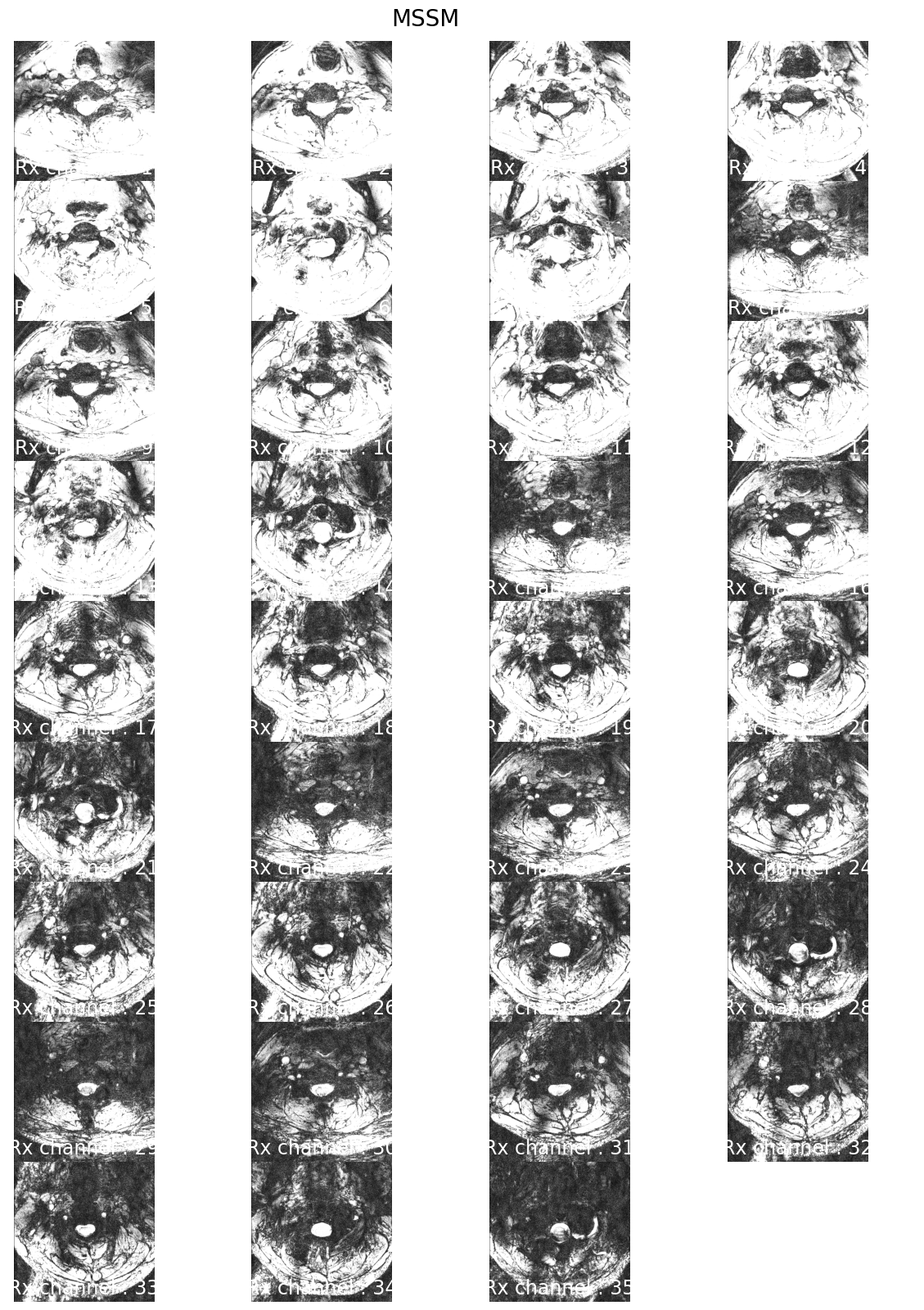
Finished#
# Indicate duration of data processing
end_time = datetime.now()
total_time = (end_time - start_time).total_seconds()
# Convert seconds to a timedelta object
total_time_delta = timedelta(seconds=total_time)
# Format the timedelta object to a string
formatted_time = str(total_time_delta)
# Pad the string representation if less than an hour
formatted_time = formatted_time.rjust(8, '0')
print(f"Total Runtime [hour:min:sec]: {formatted_time}")
Total Runtime [hour:min:sec]: 1:23:38.777888
